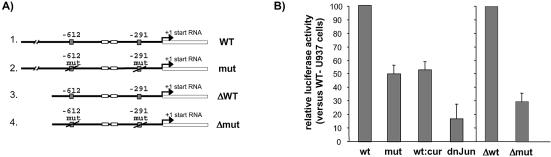
Figure 6.
TPA-enhancement of the hMSH2 promoter is dependent on functional AP-1 binding sites. U937 cells were transiently cotransfected with derivatives of luciferase reporter vectors according to the procedure outlined in Materials and Methods, and afterwards treated or not by TPA (100 nM for 6 h), as indicated. (A) A schematic structure of the constructs in pGL3 plasmid represents the luciferase gene (open box) fused to various forms of the hMSH2 promoter sequence (bold line). A 1.88 kb fragment of the hMSH2 promoter region carrying the AP-1 binding sites, either wild type (wt) or mutated (mut), was inserted upstream of the luciferase gene. Constructs ‘Δwt’ and ‘Δmut’ correspond to the 5′ truncated sequence of the wild type and mutated promoter region of hMSH2 due to a 930 bp deletion. (B) TPA-stimulated U937 cells were transfected with either wild type or mut hMSH2 promoter, or with the deleted forms of the plasmids. ‘Wt:cur’ means that U937 cells transfected with the wild type hMSH2 promoter were treated with curcumin (50 µM) for 1 h prior to addition of TPA. For ‘dn-Jun’, transfections with wild type hMSH2 promoter were performed on TPA-treated U937 cells containing the dominant negative mutant of c-Jun. Results are shown as the relative luciferase activity reported to that of TPA-treated U937 transfected with the wild type hMSH2 promoter, except for transfections with the Δmut deleted plasmid where the ratio is normalised to transfections with the Δwt plasmid. Data are the means of three independent experiments.