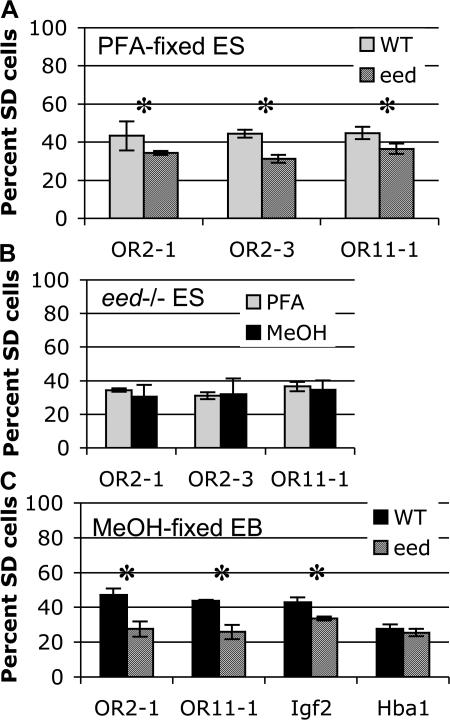
Figure 4.
Eed is required for SIAR. (A) Mutation of Eed reduces SIAR in ES cells. The frequency of SD FISH signals for three OR arrays in PFA-fixed wild-type (gray) and Eed knockout (striped) ES cells is shown. Asterisks indicate a significant difference between wild-type and Eed as determined by an unpaired t test (P < 0.005). Error bars represent one standard deviation in each direction. See Fig. S4 for complete scoring of SS, SD, and DD signals (available at http://www.jcb.org/cgi/content/full/jcb.200706053/DC1). (B) PFA-fixed and MeOH-fixed Eed −/− ES cells display a similar frequency of SD signals. The frequency of SD FISH signals for three OR arrays in PFA-fixed (gray) and MeOH-fixed (black) Eed knockout ES cells is shown. Error bars represent one standard deviation in each direction. See Fig. S4 for complete scoring of SS, SD, and DD signals. (C) Asynchronous replication of random monoallelic loci is lost in differentiated Eed −/− cells. The frequency of SD FISH signals for two OR arrays, Igf2, and Hba1 in MeOH-fixed wild-type (black) and Eed knockout (striped) differentiated cells is shown. EBs were differentiated for 17 d, and differentiation was tested using an alkaline phosphatase assay. 97.5% of wild-type cells and 95% of Eed −/− cells were negative for phosphatase activity. Asterisks indicate a significant difference between wild-type and Eed as determined by an unpaired t test (P < 0.001). Error bars represent one standard deviation in each direction. See Fig. S4 for complete scoring of SS, SD, and DD signals.