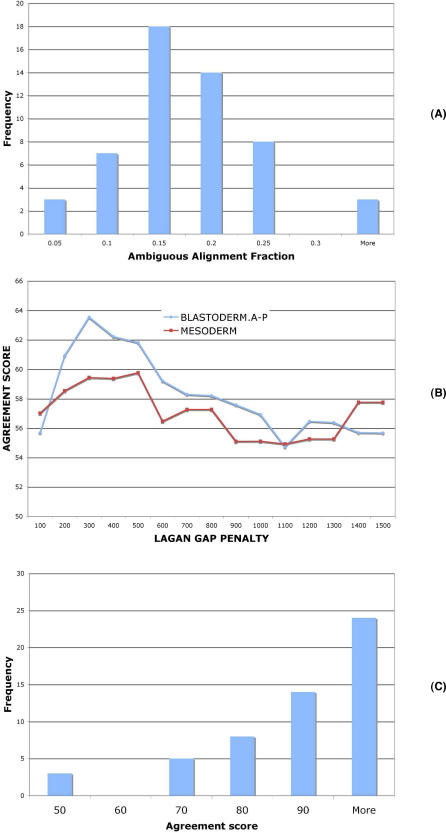
Figure 4. Morphalign Alignments: Differences from Alternative Alignment Methods.
(A) For each CRM in the BLASTODERM.A-P set, the fraction of the D. melanogaster sequence that was ambiguously aligned (to two or more positions in D. mojavensis) was computed. The figure shows the histogram of these alignment ambiguity fractions for the set.
(B) Median of alignment agreement scores between output of Morphalign and output of LAGAN (run separately with a range of gap opening penalties) for CRMs in the BLASTODERM.A-P (blue) and the MESODERM (red) sets.
(C) Histogram of alignment agreement scores between Morphalign and its no-motifs version, for the BLASTODERM.A-P set of CRMs.