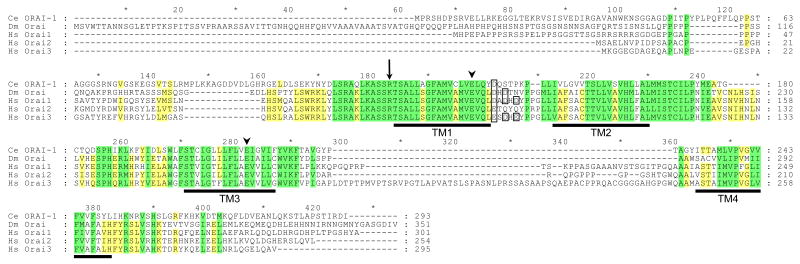
Figure 5.
Amino acid sequence alignment of C. elegans (Ce) ORAI-1 with Drosophila (Dm) Orai and human (Hs) Orai1, Orai2 and Orai3. Yellow and green shading indicates sequence identity and conserved amino acid substitutions, respectively. Transmembrane (TM) domains of ORAI-1 predicted by TMpred (http://www.ch.embnet.org/software/TMPRED_form.html#) are underlined in black. A conserved arginine residue mutated in SCID patients lacking lymphocyte CRAC activity is denoted by the arrow. Conserved glutamate residues located in TM1 and TM3 that contribute to channel ionic selectivity are denoted by arrowheads. Alignment was performed using Vector NTI software (InforMax, Bethesda, MD). Percent identity and similarity of C. elegans ORAI-1 to Drosophila Orai, human Orai1, human Orai2 and human Orai3 were 34% and 55%, 34% and 54%, 38% and 59%, and 35% and 59%, respectively. Acidic residues in the extracellular loop between TM1 and TM2 that may control channel selectivity are outlined in black.