Abstract
Our objective is to identify genes regulating metastasis of osteogenic sarcoma (OGS) since metastasis is the primary cause of mortality among patients with OGS. To identify such genes, we first created a database of differentially expressed genes between six low-grade and six high-grade OGS tumors, and between a normal immortalized osteoblast cell line (FOB) and four commercially available OGS-derived cell lines. We specifically searched for surface-proteins over-expressed in high-grade OGS, since we hypothesize that tumor-cell specific surface markers are key to metastasis. A gene encoding Tumor Endothelial Marker7 (TEM7) was selected as a candidate for further study. TEM7 expression pattern was assessed by RT-PCR, Western blotting and immunostaining. TEM7 mRNA was abundantly expressed in SAOS cells (derived from high-grade OGS), but not in FOB or MG63 cells (derived from low-grade OGS). Virtually no expression of TEM7 protein was observed in FOB cells but abundant expression was noted in SAOS and TE85 cells. Employing immunostaining of 92 human OGS specimens (50 high grade and 42 low-grade) collected before chemotherapy show 97% (37 of 38) of high-grade OGS specimens with metastasis have high TEM7 staining. Further, we found that elevated expression of TEM7 correlated with poor survival (p<0.04) of affected patients. Inhibiting TEM7 function by siRNA inhibited invasion and migration of OGS cells with metastatic potential. Our results suggest TEM7 expression level closely parallels histology-based prognostication of OGS metastasis and, therefore, it is a therapeutic target. This is the first demonstration of a link between TEM7 and cancer metastasis.
Keywords: TEM7, Osteogenic sarcoma, Metastasis, siRNA, Tumor marker
1. introduction
Osteogenic sarcoma (OGS) is the most common primary malignancy of bone in the pediatric age group, with a peak incidence of 400 cases in the second decade of life each year in the US (Gurney et al., 1999). The introduction of multiagent chemotherapy in addition to surgery has dramatically increased long-term survival for patients with OGS, with a disease-free 5-year survival reaching 65–70% for patients with localized disease (Link et al., 1986; Meyers and Gorlick, 1997; Arndt and Crist, 1999; Hawkins and Arndt, 2003). However, patients who develop metastatic disease still have an extremely dismal prognosis despite its contemporary treatment. Recently, the 4-year disease free survival for patients with metastatic disease was reported to be only 6% (Hawkins and Arndt, 2003). Therefore, new treatment strategies are needed. Metastases are the cause of 90% of cancer deaths for patients with OGS (Hanahan and Weinberg, 2000). Metastasis is a complex and multistage process involving local invasion, access to the circulation, seeding and eventual proliferation within a favorable distant target organ (Pantel and Otte, 2001; Fidler, 2002b; Bogenrieder and Herlyn, 2003). A few new paradigms such as metastasis suppressors (Shevde and Welch, 2003; Steeg, 2003), and scatter factors have emerged recently by analyzing this process at the molecular level, (Trusolino and Comoglio, 2002; Birchmeier et al., 2003; Zhang and Vande Woude, 2003). Despite these recent advances, the process of metastasis still remains incompletely characterized at both the molecular and biochemical levels (Fidler, 2002a).
High grade is the best available prognostic indicator of OGS metastasis, which, in turn, is the most informative indicator for poor survival. Grade of an OGS specimen is determined histologically, which is a sum-total of diverse molecular processes. Molecular factors that determine tumor grade are of great academic and clinical relevance because a marker-based approach can provide a uniform approach to categorizing tumors (grade etc.), and the markers themselves can be considered as rational targets for developing novel therapy. Our long-term goal is to identify genes able to predict metastasis and/or survival outcome of patients with OGS.
A rational approach for identifying molecular targets for metastasis should combine in vitro manipulation followed by eventual validation using clinical specimens. We searched for candidate metastasis-associated genes in our database of differentially expressed genes in OGS, which was created by comparing gene expression profiles between cell lines derived from low-grade and high-grade OGS (Fuchs et al., 2000a; Fuchs et al., 2000b) and between low-grade and high-grade tumor specimens (our unpublished data). High-grade OGS usually metastasizes to lung. We specifically searched for surface-proteins over-expressed in high-grade OGS cells, since we hypothesize that tumor-cell specific surface markers are key to metastasis. We identified TEM7 for further study because 1. TEM7 mRNA was found to be over-expressed in high-grade OGS-derived specimens in our unpublished database; 2. TEM7 mRNA expression has been reported to be elevated in endothelial cells derived from tumors (St Croix et al., 2000; Carson-Walter et al., 2001; Nanda and St Croix, 2004), and 3. TEM7 has been reported to be a membrane –associated protein (St Croix et al., 2000; Nanda and St Croix, 2004). Additionally, our observation of high expression of TEM7 mRNA in OGS cells, which are of mesenchymal origin was considered to be intriguing. Taken together, we inferred that TEM7 is an important gene associated with high-grade OGS and, consequently, with metastasis. Most recently, using a comparable approach, Khanna et al reported that the membrane-cytoskeleton linker ezrin, a member of the Band 4.1 superfamily of ERM (ezrin-radixin-moesin) proteins, is necessary for osteogenic sarcoma metastasis, and that there is a significant association between high ezrin expression and poor outcome in pediatric osteosarcoma patients (Khanna et al., 2004). However, we suspect that ezrin is not the only protein regulating OGS metastasis, and recently Kang et al. have shown that a set of four genes determine breast cancer metastasis to bone (Kang et al., 2003).
In this report, we confirmed TEM7 expression in OGS tumor cells by RT-PCR and Western blotting, and evaluated the expression pattern of TEM7 in OGS specimens by immunostaining. Further, we developed siRNA-based approach to inhibit TEM7 expression and subsequently evaluated various key cellular steps leading to metastasis using cell-based assays. Our results suggest that TEM7 has a regulatory role in multiple steps necessary for OGS metastasis.
2. Materials and methods
2.1. Cell Culture
MG63, TE85, SaOS and U2OS cell lines were obtained from ATCC, and the fetal osteoblast (FOB) cell line was a kind gift from Dr. TC. Spelsberg at the Mayo Clinic. The cell lines were maintained in DMEM/F12 with 10% FCS at 37 ºC and 5 % incubator. A monoclonal antibody against TEM7 (IMG-402) was purchased from Imgenex, San Diego, CA.
2.2. Isolation of RNA
Cytoplasmic RNA was isolated from cells as indicated and grown on 6 well plates. Cells were washed with PBS then lysed in Trizol (GIBCO-BRL) according to the manufacturer’s protocol. RNA samples were treated with DNase to destroy contaminating genomic DNA (Sanyal et al., 1997). A total of 2μg of total RNA from each sample was converted to cDNA.
2.3. Reverse Transcriptase Polymerase Chain Reaction for TEM7 mRNA
The RT-PCR was essentially performed as described by Fuchs et al. (Fuchs et al., 2000b). Briefly, RT-PCR was performed in a total volume of 20 uL containing 10 mM Tri-HCl, pH 8.3, 200 uM of each dNTP, 1.5 mM MgCl2, 2 uM of each primer for TEM7 mRNA, 2 uL of randomly primed cDNA, and 0.5 U of Amplitaq. Primers from desired segments of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA were included. PCR were performed by first heating the reaction at 94°C for 5 min, then cycling the reaction 35 times at 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min. Amplification products (5uL) were analyzed by electrophoresis on 2% agarose gel, which was stained with ethidium bromide for visualizing DNA.
2.4. Western Blotting
Total protein was extracted from the indicated cells with lysis buffer (0.15MNaCl, 5mM EDTA, pH8, 10mM Tris-Cl, pH7.4, 1% Triton-X100). Protein concentrations were determined by Bradford assay. Equal amounts of protein (20μg) were electrophoresed by 10% SDS-PAGE and transferred onto a nylon membrane (Millipore Corporation). The membrane was incubated with antibodies to TEM7 (IMGENEX, SanDiego) at a dilution of 1:1000, followed by incubation with peroxidase-linked species-specific whole antibody (ECL Anti-mouse IgG) at a dilution of 1:30,000 (Amersham, Biosciences), and then developed with ECL reagent (Amersham Biosciences).
2.5. Generation of synthetic siRNA
The two si-RNA sequences targeting the human TEM7 chosen in this study (from mRNA sequence; GenBank™ accession number AF279144) corresponds to sequences 839 – 861 (5′AAAACCGGCCUAUCGGAUGCC-3′), and 894–916 (5′AATCTCGGCGAAGGAGCATCT–3′), respectively. The si-RNAs were 21 nucleotides long with symmetric 2-nucleotide 3′ overhangs composed of 2′-deoxythymidine to enhance nuclease resistance. The siRNAs were synthesized chemically with standard purification (Ambion, Austin, Texas). Scrambled siRNA was also obtained from Ambion. Sense RNA sequence for the first target was 5′AACCGGCCUAUCGGAUGCCTT-3′, and antisense RNA 5′GGCAUCCGAUAGGCCGGUUTT3′ whereas for the second target the sense RNA sequence was 5′-UCUCGGCGAAGGAGCAUCUTT-3′ and the antisense RNA sequence was 5′AGAUGCUCCUUCGCCGAGATT-3′.
2.6. Transfection of cells with siRNA
Before transfection, cells were cultured at 50–70% confluence in 6-well plates (10 cm2) and were washed two times with PBS. Then the cells were transfected with duplex siRNA using GeneSilencer™ siRNA transfection reagent (Gene Therapy Systems, San Diego) according to the manufacturer’s instructions. Unless otherwise described, cells were transfected with100 nM siRNA duplex in 1 ml of transfection medium without FCS for 4–6 h after which the medium was changed and adjusted to 1.5 ml per well with DMEM +10% FCS. Silencer™ GAPDH negative control siRNA (Ambion, San Diego) was used as negative control under similar conditions (100 nM).
2.7. Migration and invasion in presence of siRNA against TEM7
SaOS, TE85 and MG63 osteosarcoma cell lines (in presence and absence of the siRNAs) were used for all these assays. All these assays were performed with commercially available kits using the protocol supplied by the manufacturer. For evaluating cell migration, first cell density was adjusted to 1 × 106 cells/ml in 10% DMEM, 100,000 cells in 100 μl were added to the top chamber (Boyden chamber) of a 96 well plate apparatus (Chemicon International, CA), using a 6.5-mm diameter, 8-μm pore size in the absence or presence of 10% FCS in the lower chamber. After overnight incubation at 37°C in an atmosphere containing 5% CO2, live cells that passed through the membrane were collected from the lower well and lysed in a CyQuant GR International, CA). Invasion was evaluated as above in 96 well plates containing inserts of 8μm pore size polycarbonate membrane coated with a thin layer of ECMatrix ™. Results from triplicate wells were expressed as mean +−S.D. All experiments were repeated at least once.
2.8. Immunostaining
Human paraffin-embedded osteosarcoma tissues (prior to chemotherapy) were cut in 5 μm-thick sections and placed on superfrost charged slides. The slides were deparaffinized in xylene (2 × 5 min), successively soaked in absolute ethanol and a series of ethanol/water mixtures, and finally rinsed with tap water. Endogenous peroxidase activity was blocked by using 0.3% H2O2 in methanol (1:1 vol). After a tap water rinse, sections were placed into 1 mM pre-heated (~90°C) EDTA (pH 8) and then steamed for 30 min. After cooling in buffered 1 mM EDTA (pH 8) for 5 min, the sections were rinsed in tap water and placed in phosphate-buffered saline (pH 7.4). Sections were blocked for 5 min with DAKO Peroxidase Block (DAKO Cytomation, Carpenteria, CA) and then incubated with the primary anti-TEM7 antibody (IMGENEX, San Diego) at a 1:10,000 dilution for 30 min. The DAKO mouse EnVision+ HRP System/AEC+ and the DAKO Autostainer were utilized for detection. Sections were counterstained with light hematoxylin and then mounted with a coverslip. Basic routine Hematoxylin and Eosin staining (modified Schmidt’s hematoxylin) was performed for all specimens to ensure tissue quality.
2.9. Grading of TEM7 immunostaining
TEM7 staining was graded according to its intensity from 0 to 4. (0 = no staining; 1 = some (<25%) islands of staining; 2 = homogenous weak staining over 25%–50% of the slide; 3 = intermediate intensity and/or islands of intense staining covering 50%–75% of the slide; 4 = intense homogeneous staining covering >75% of the slide. The study was approved by the institutional review board.
2.8. Statistical Analysis
Data were evaluated through ANOVA. A significance level of 0.05 was used for all tests.
3. Results
3.1. Cells derived from high-grade OGS express high levels of TEM7 mRNA and protein
Differential expression of TEM7 mRNA in OGS was first observed in our laboratory when mRNA expression profile from OGS-derived cells was compared with that of the FOB cells by preferential amplification of coding sequences (PACS) (Fuchs et al., 2000a; Fuchs et al., 2000b). To validate this observation, RT-PCR was performed on total RNA from FOB, MG63 and SAOS cells, a representative result from these experiments is presented in Figure 1A. As could be seen, the level of RT-PCR product corresponding to TEM7 mRNA appears to be much more in SAOS cells than in FOB or MG63 cells.
Figure 1. TEM7 expression in OGS cells evaluated by RT-PCR (A)Western blotting (B).
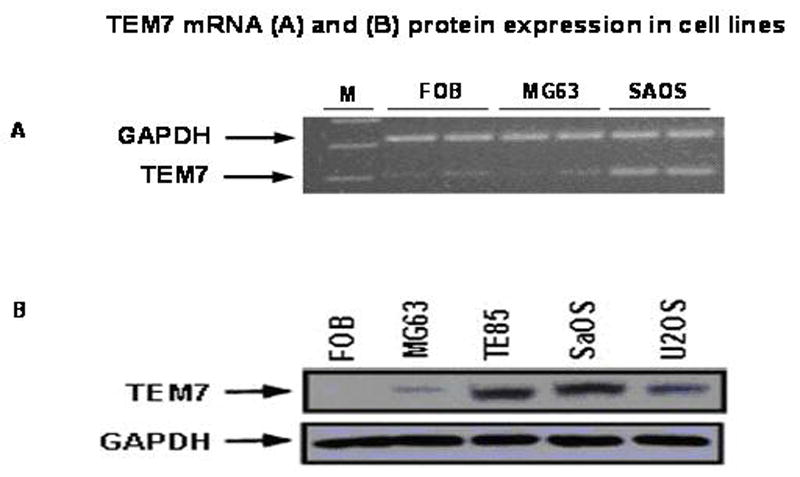
Cell lines are marked. FOB is an immortalized normal osteoblast cell line. For western blotting, the same blot was successively probed withTEM7 and GAPDH (glyceraldehydes acid 3-phosphate dehydrogenase) antibodies, the latter to primarily serve as control for protein loading. Primers used for RT-PCR are 5′CTGCCACCCAGAAGACTGTGGAT3′ and CGCTGTTGAAGTCAGAGGAGACC3′ for GAPDH mRNA (from accession number M33197), and 5′GACGGCCTTCAGAACAACCTGTC3′ and 5′GGTGGCTGCGAAACTTCATGGCT3′ for TEM7 mRNA (from accession number AF 279144). The GAPDH primers amplify an ~320 bp band and the TEM7 primers amplify a band of 207bp. (Based on alignment with human genomic sequence, it appears that the TEM7 primers should amplify a band of 2102 bp from genomic DNA, which, based on our experience, is never amplified from a cDNA preparation).
To evaluate TEM7 expression at the protein level, total cell homogenate from OGS-derived cell lines was subjected to analysis by Western blotting with an antibody raised against human TEM7. A representative result from these experiments, presented in Fig 1B, show no expression of TEM7 protein in FOB cells, very little in MG63 cells but a high expression level in SaOS, U2OS and TE85 cells. The antibody identified a band of ~57Kd. These results provide evidence for a high level expression of both TEM7 mRNA and protein in OGS-derived cells. TEM7 was originally shown to be expressed by (tumor) cells of endothelial origin (Carson-Walter et al., 2001; Nanda and St Croix, 2004), thus TEM7 expression by osteogenic sarcoma-derived cells, which are cells of mesenchymal origin (note that non-tumorigenic bone cells such as FOB, although of mesenchymal origin, does not express TEM7) was considered intriguing and provided a basis for further study. Nine of eighteen primary OGS tumor specimens were also found to express TEM7 mRNA (data not shown).
3.2. High-grade OGS tumor specimens have an elevated level of TEM7 expression
To investigate the above putative relationship further, we determined TEM7 expression by immunostaining of OGS specimens for which the clinical outcome is known. We performed immunohistochemical analyses to assess TEM7 protein expression in 92 tumor specimens (44 metastatic, 48 non-metastatic) from patients with OGS. This cohort of patients had roughly an equal distribution with high-grade and low-grade lesions (50 and 42, respectively). A concise description of the OGS specimens studied, including TEM7 staining patterns, metastasis and survival outcome is presented in Table 1. Overall, 54% of the tumors are of high grade and 46% low grade. 37 specimens with staining intensity between 0 –2 were from low-grade OGS, and 48 samples with staining intensity between 3–4 were from high-grade OGS. Thus TEM7 appears to be predominantly expressed in high-grade OGS. Indeed, a statistically significant relationship between TEM7 expression and high-grade OGS was observed (p<0.001).
Table 1. Relationship between TEM7 immunostaining with tumor grade, metastasis and survival of patient with OGS.
Clinical history data and TEM7 staining intensity of all the 92 samples are presented. The successive numbers in each box were generated manually. 1 – Grades 1 and 2 are low-grade and grades 3 and 4 are considered high-grade tumors. 2 – TEM7 staining grades (see Materials and Methods) 3 to 4 are considered high-grade staining; TEM7 staining of 0–2 are considered low-grade staining. 3 – Survival greater than 60 months has been considered ‘good’; survival of 60 months or less has been considered ‘poor’.
| Categories | High grade OGS1 50 | Low grade OGS1 42 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metastasis | 38 | 6 | |||||||||||||||
| No metastasis | 12 | 36 | |||||||||||||||
| TEM 7 Staining | High2 | 37 | 11 | 1 | 4 | ||||||||||||
| Low2 | 1 | 1 | 5 | 32 | |||||||||||||
| Survival outcome | Good3 | 9 | 0 | 7 | 1 | 1 | 4 | 4 | 29 | ||||||||
| Poor3 | 28 | 1 | 4 | 0 | 0 | 1 | 0 | 3 | |||||||||
Since metastatic disease is common in high-grade OGS, we intended to evaluate if there is any relationship between TEM7 expression and metastasis. We found a significantly higher staining in tumor specimens of patients who developed metastatic disease compared with those who didn’t. (3.0 versus 1.6; p<0.0001). Overall, 76% (38/50) of high grade tumors and 14% of low-grade tumors had metastasis. 97% (37/38) of high grade tumors with metastasis had high TEM7 staining. Altogether, these data suggest that TEM7 is involved in disease progression in patients with osteogenic sarcoma.
3.3. High TEM7 expression is comparable with high grade as a prognostic indicator of poor survival of patients with OGS
We observed a strong relationship between high grade OGS and high TEM7 expression and between high TEM7 expression and metastasis. Since metastasis results in poor survival of patients, we hypothesized that high TEM7 expression is correlated with poor survival of patients with OGS. To test this hypothesis, we first collected the survival record of 92 patients with OGS from the Mayo Clinic database and the data analyzed. Since tumor grade (high) is a poor prognosticator of survival of patients with OGS, we wanted to evaluate if high TEM7 staining alone can predict survival comparable to high grade alone. Results of these analyses are presented in Table 1 and Figure 2. We find that 66% (33/50) of patients with high-grade tumor have poor survival (p<0.03), whereas 60% (28+4)/(37+11+1+4) of patients with tumors having high TEM7 expression (irrespective of grade) have poor survival (p<0.04, also see Table 1). We conclude that although high grade is a better predictor of patient survival compared to TEM7 staining (p<0.03 vs. p<0.04), high TEM7 staining compares well with high grade as a prognosticator of patient survival from OGS. It is interesting to note that 4/4 low-grade tumors with high TEM7 expression had good survival, suggesting factors beyond TEM7 as regulating the complex process of survival from the disease.
Figure 2. TEM7 expression in OGS specimens and survival analysis.
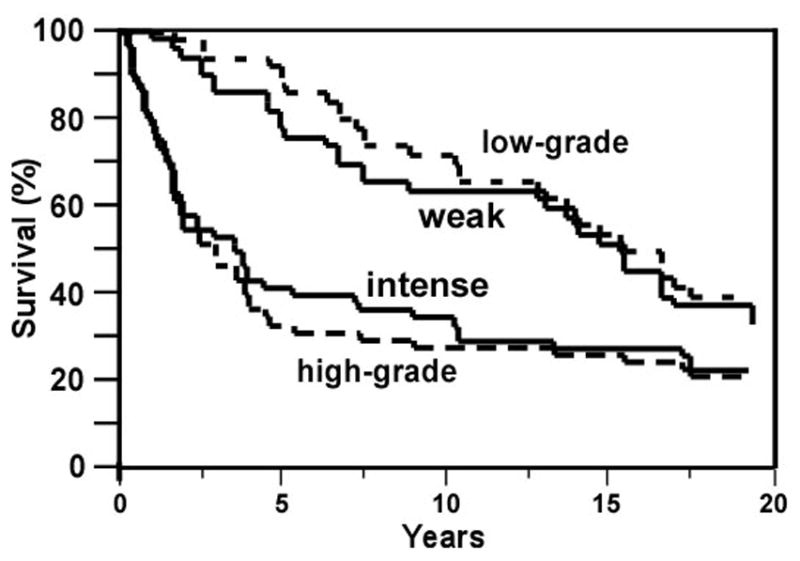
Kaplan-Meier curve comparing the survival of patients with weak staining (grade 0 to 2, marked as ‘weak’) and intense staining for TEM7 (grade 3 to 4, marked as ‘intense’; p<0.04), and with low grade (grade 0 to 2) and high-grade (grade 3 to 4; p<0.03).
3.4. siRNAs directed against TEM7 inhibit expression of TEM7 in osteogenic sarcoma cells
Our findings based on cell lines and tumor tissue analyses presented above imply a potentially important role of TEM7 in osteogenic sarcoma. To explore any specific role of TEM7 in any aspect of osteogenic sarcoma, the first step is to develop a cellular model that sustains forced inhibition of TEM7 expression. Based on this rationale, we employed siRNA to silence expression of TEM7 in SaOS and TE85 cells, which both showed high level of expression of TEM7. We employed two different siRNA oligos, which alone or in combination efficiently knocked-down TEM7 expression in both cell lines (Fig. 3). It appears that the siRNAs were active at both time points tested, however, inhibition of TEM7 expression was stronger at 48h than at 72h – the underlying reason for this was not explored further (scrambled siRNA obtained from Ambion did not interfere with expression of either TEM7 or GAPDH – data not shown). We concluded from the results of these experiments that equimolar mixture of the two siRNAs would produce maximal effects on relevant cellular function.
Figure 3. Silencing of TEM7 expression in TE85 and SaOS cells by siRNA.

Two different siRNAs (TEM7-1 and TEM7-2) were used alone or in combination. Cell homogenate was prepared after indicated time of transfection with siRNA and 20 ug of homogenate was subjected to Western blotting with TEM7 and GAPDH antibodies successively. A – 48 hours after transfection. B – 72 hours after transfection. The scrambled siRNA was used only for the 72h experiment.
3.5 Inhibiting TEM7 expression by siRNA inhibits migration and invasion of TE85 and SAOS cells in vitro
Having established the conditions for silencing TEM7 expression in cultured cells, we used these cell lines to assess the role of TEM7 on migration and invasion of various cells. These steps are necessary for successful metastasis by a cancer cell. We first tested TEM7-silenced and non-silenced osteogenic sarcoma cell lines for the ability of migration. Our data showed that silencing of TEM7 in both TE85 and SaOS cell lines was followed by a significantly reduced ability (~50% reduction) of the cells to migrate (Fig. 4, Top panel). We then employed the same system to test for any role of TEM7 in matrix invasion, another hallmark of metastasis. We found that silencing of TEM7 had a profound negative impact on invasion in SaOS cells (~50% reduction), but not in TE85 cells (Figure 4, bottom panel). These data imply that cells expressing a high level of TEM7 depend on this protein for invasion and migration than cells expressing a low level or lacking TEM7.
Figure 4. Evaluation of cell migration, invasion and proliferation with respect to TEM7 expression.
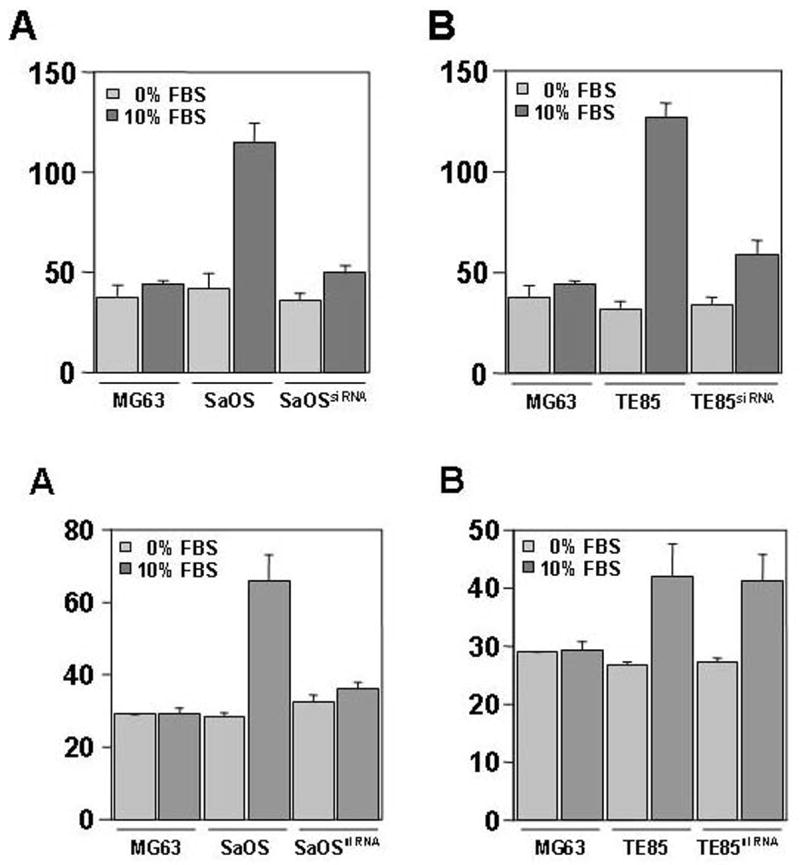
Top: Cell migration studies - The low-grade MG63 cells (express very little TEM7) were used for comparison of cell-migration, these cells did not receive any siRNA. Cells were grown in presence of 0% (light bars) and 10% FBS (dark bars). A – migration of SaOS cells; B – migration of TE85 cells. Both SaOS and TE85 cells show drastic inhibition of migration in presence of TEM7 siRNA. In these experiments, both siRNAs were used together. Middle: Invasion studies : A- invasion by SaOS cells; B – invasion by TE85 cells. Invasion by SaOS cells are more affected under TEM7 silencing than in TE85 cells. Bottom: Cell proliferation studies - Cells were grown in presence of 10% FBS. Both siRNAs, together, were used. A ~30% reduction in proliferation is observed for SaOS and TE85 cells, however, the siRNAs had no effect on the proliferation of MG63 cells.
4. Discussion
Molecular markers that regulate initiation and propagation of the metastatic process of osteogenic sarcoma are largely unknown, therefore, prognostication and therapeutic course of patients with OGS is largely guided by tumor grade. Clearly, identification of molecular factors that govern metastasis and/or other clinical parameter will have a significant implication in future management of patients with OGS. By combining a global and candidate molecular marker (CMM)-based screening strategy between non-aggressive and aggressive OGS tumors and cell lines, we identified TEM7 as a candidate gene associated with high grade (therefore, potentially with metastasis) in osteogenic sarcoma. Since TEM7 was preferentially up-regulated in cell lines originating from aggressive tumors both at the mRNA and protein levels, we extended our analysis by immunohistochemistry on a large number of patient specimens. These analyses uncovered a potential relation between various clinical outcomes (tumor grade, metastatic potential and survival) of patients with OGS and increased TEM7 expression. We further show that silencing of TEM7 expression in OGS- derived cell lines has considerable inhibitory effect on invasion, migration and proliferation of the cells – key steps necessary for metastasis. Whether a common signaling cascade regulated by TEM7 underlies the inhibitory effect on these steps remains to be elucidated.
The family of TEM proteins was discovered as a result of an expanded search for specifically targeting the endothelium of tumor blood vessels as a new anticancer regimen ( St Croix et al., 2000). TEM proteins are believed to be located on the cell surface; therefore, they may be of particular value as therapeutic targets because of a relatively easy access to a drug. Together with TEM7R (TEM7-related, which encodes a protein having 57% amino acid identity to TEM7), TEM7 belongs to a new family of cell surface proteins. These two proteins have a single pass transmembrane protein with a conserved cytoplasmic tail, which is, however, unrelated to other proteins (Carson-Walter et al., 2001; Nanda and St Croix, 2004). They contain a plexin-like as well as a weak nidogen-like domain, which may be involved in mediating protein-protein interactions (Fogh and Trempe, 1975).
Although the initial report (St Croix et al., 2000; Carson-Walter et al., 2001) did not exclude the possibility of TEM7 expression in cells other than endothelial cells of tumor origin, we consider it intriguing that TEM7 is expressed in osteogenic sarcoma cells, which is of mesenchymal origin. Indeed, there is some indication in the literature that implies that osteogenic sarcoma cells may have the capacity to adopt an endothelial cell phenotype. Khanna et al. reported strong evidence for this theory by assessing metastasis-associated differences in a murine-model of osteosarcoma. These investigators have shown that osteosarcoma cells express aberrant endothelial- and other vascular-associated markers such as CD31, TIE-1, TIE2, and FLT-1, FLT-4 (Khanna et al., 2001). Similar to our findings in this report, it may be not surprising that such endothelial markers were found to be associated clinically with metastasis (Khanna et al., 2001). Indeed, as evident from our study, it is possible that tumor cells, which express endothelial markers, may be clinically aggressive in particular, and render a tumor lesion metastatic.
While there is no clear answer to why a tumor cell should express endothelial markers, it is possible that such a phenotype contribute to “vascular mimicry” (Maniotis et al., 1999). Vasculogenic mimicry has been described as the ability of a tumor cell to express endothelium-associated genes and form extracellular matrix rich in vasculogenic networks in three-dimensional culture, thereby recapitulating embryogenic vasculogenesis. These networks may serve as an intratumoral fluid-conducting network (Hendrix et al., 2003). It has recently been shown that vascular mimicry may also be functional in osteosarcoma. Cai et al. have shown that U2OS cells generate vascular channels, which facilitate tumor perfusion independent of tumor angiogenesis (Cai et al., 2004). The molecular mechanism underlying vascular mimicry or neoangiogenesis, especially in the context of OGS is not known. Although, based on our initial set of experiments, we failed to obtain a positive correlation between TEM7 expression and tube formation in vitro (data not shown), TEM7 may still contribute to neovasculogenesis in other steps since this process involves multiple steps. Indeed, since we find an inhibitory effect of TEM7 siRNA on cell proliferation and migration (Figure 3), TEM7 might have an indirect role on angiogenesis because proliferation and migration are critical steps for angiogenesis.
The only other protein that has been demonstrated to have a role in OGS metastasis is ezrin (Khanna et al., 2004). However, our data (not shown) on the pattern of ezrin expression (all OGS-derived cells tested including cells that are known to be non-tumorigenic and normal immortalized osteoblast show both ezrin mRNA and protein expression; although cells that create metastasis in animal model do have higher ezrin expression than cells that do not create metastasis) suggest that ezrin alone is not sufficient to cause OGS metastasis. We speculate that ezrin may cross-talk with TEM7 and that neither molecule alone is sufficient for OGS metastasis. Since TEM7 is a transmembrane protein (it has recently been reported that there are, in addition to the membrane-bound form, soluble and secreted forms of TEM7 (Nanda et al., 2004) and since ezrin provides a link between membrane and the cytoskeleton, it is possible that extracellular signals pertinent to OGS metastasis is first recognized by TEM7 (or similar molecule) and then transferred to the cytoskeleton through ezrin (or similar molecule). Evidently, such a concept will need to be evaluated through an appropriate animal model.
Acknowledgments
This work was supported by a career development award (to B. F.) from the European Federation of Orthopedics and Traumatology (EFORT), and Zimmer, and by a grant (AR47974) from the National Institutes of Health (NIH). We thank Dr. S. Barik (University of South Alabama) for designing the siRNAs.
The abbreviations used are
- TEM7
tumor endothelial marker 7
- PACS
preferential amplification of coding sequences
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arndt CA, Crist WM. Common musculoskeletal tumors of childhood and adolescence. New England Journal of Medicine. 1999;341:342–2. doi: 10.1056/NEJM199907293410507. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nature Reviews Molecular Cell Biology. 2003;4:915–5. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Bogenrieder T, Herlyn M. Axis of evil: molecular mechanisms of cancer metastasis. Oncogene. 2003;22:6524–6. doi: 10.1038/sj.onc.1206757. [DOI] [PubMed] [Google Scholar]
- Cai XS, Jia YW, Mei J, Tang RY. Tumor blood vessels formation in osteosarcoma: vasculogenesis mimicry. Chinese Medical Journal. 2004;117:94–8. [PubMed] [Google Scholar]
- Carson-Walter EB, Watkins DN, Nanda A, Vogelstein B, Kinzler KW, St Croix B. Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Research. 2001;61:6649–55. [PubMed] [Google Scholar]
- Fidler IJ. Critical determinants of metastasis. Seminars in Cancer Biology. 2002a;12:89–96. doi: 10.1006/scbi.2001.0416. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The organ microenvironment and cancer metastasis. Differentiation. 2002b;70:498–505. doi: 10.1046/j.1432-0436.2002.700904.x. [DOI] [PubMed] [Google Scholar]
- Fogh J, Trempe G. In: Human Tumor Cells In Vitro. Fogh J, editor. Plenum Press; New York: 1975. pp. 115–159. [Google Scholar]
- Fuchs B, Zhang K, Bolander ME, Sarkar G. Differential mRNA fingerprinting by preferential amplification of coding sequences. Gene. 2000a;258:155–63. doi: 10.1016/s0378-1119(00)00393-0. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Zhang K, Bolander ME, Sarkar G. Identification of differentially expressed genes by mutually subtracted RNA fingerprinting. Analytical Biochemistry. 2000b;286:91–8. doi: 10.1006/abio.2000.4792. [DOI] [PubMed] [Google Scholar]
- Gurney J, Swensen A, Bulterys M. Cancer Incidence and Survival among Children and Adolescents: United States SEER program 1975–1995. NIH; Bethesda: 1999. [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hawkins DS, Arndt CA. Pattern of disease recurrence and prognostic factors in patients with osteosarcoma treated with contemporary chemotherapy. Cancer. 2003;98:2447–56. doi: 10.1002/cncr.11799. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Vasculogenic mimicry and tumour-cell plasticity: lessons from melanoma. Nature Reviews Cancer. 2003;3:411–21. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–49. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Khanna C, Khan J, Nguyen P, Prehn J, Caylor J, Yeung C, Trepel J, Meltzer P, Helman L. Metastasis-associated differences in gene expression in a murine model of osteosarcoma. Cancer Research. 2001;61:3750–9. [PubMed] [Google Scholar]
- Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, Yeung C, Gorlick R, Hewitt SM, Helman LJ. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nature Medicine. 2004;10:182–6. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- Link MP, et al. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. New England Journal of Medicine. 1986;314:1600–6. doi: 10.1056/NEJM198606193142502. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe’er J, Trent JM, Meltzer PS, Hendrix MJ. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. American Journal of Pathology. 1999;155:739–52. doi: 10.1016/S0002-9440(10)65173-5. [see comment] [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister RM, Gardner MB, Greene AE, Bradt C, Nichols WW, Landing BH. Cultivation in vitro of cells derived from a human osteosarcoma. Cancer. 1971;27:397–402. doi: 10.1002/1097-0142(197102)27:2<397::aid-cncr2820270224>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Meyers PA, Gorlick R. Osteosarcoma. Pediatric Clinics of North America. 1997;44:973–89. doi: 10.1016/s0031-3955(05)70540-x. [DOI] [PubMed] [Google Scholar]
- Nanda A, et al. Identification of a binding partner for the endothelial cell surface proteins TEM7 and TEM7R. Cancer Research. 2004;64:8507–11. doi: 10.1158/0008-5472.CAN-04-2716. [DOI] [PubMed] [Google Scholar]
- Nanda A, St Croix B. Tumor endothelial markers: new targets for cancer therapy. Current Opinion in Oncology. 2004;16:44–9. doi: 10.1097/00001622-200401000-00009. [DOI] [PubMed] [Google Scholar]
- Pantel K, Otte M. Occult micrometastasis: enrichment, identification and characterization of single disseminated tumour cells. Seminars in Cancer Biology. 2001;11:327–37. doi: 10.1006/scbi.2001.0388. [DOI] [PubMed] [Google Scholar]
- Sanyal A, O’Driscoll SW, Bolander ME, Sarkar G. An effective method of completely removing contaminating genomic DNA from an RNA sample to be used for PCR. Molecular Biotechnology. 1997;8:135–7. doi: 10.1007/BF02752257. [DOI] [PubMed] [Google Scholar]
- Shevde LA, Welch DR. Metastasis suppressor pathways--an evolving paradigm. Cancer Letters. 2003;198:1–20. doi: 10.1016/s0304-3835(03)00304-5. [DOI] [PubMed] [Google Scholar]
- St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nature Reviews Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- Trusolino L, Comoglio PM. Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nature Reviews Cancer. 2002;2:289–300. doi: 10.1038/nrc779. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Vande Woude GF. HGF/SF-met signaling in the control of branching morphogenesis and invasion. Journal of Cellular Biochemistry. 2003;88:408–17. doi: 10.1002/jcb.10358. [DOI] [PubMed] [Google Scholar]


