Abstract
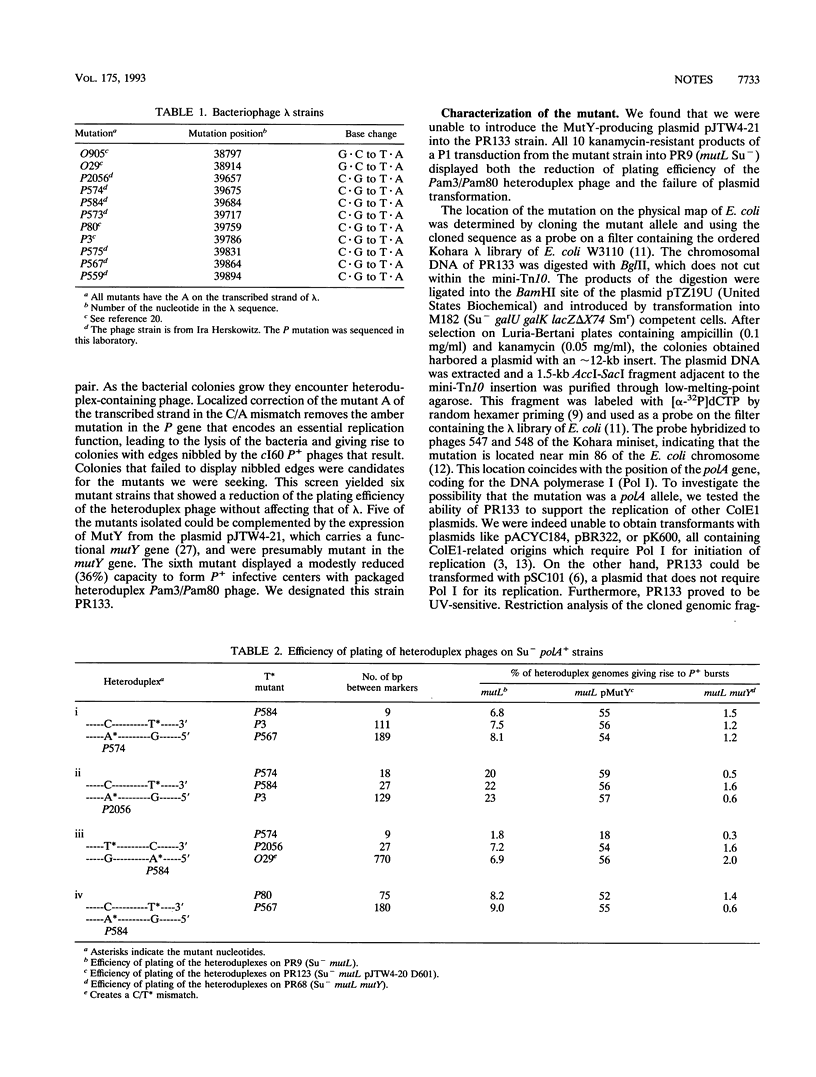
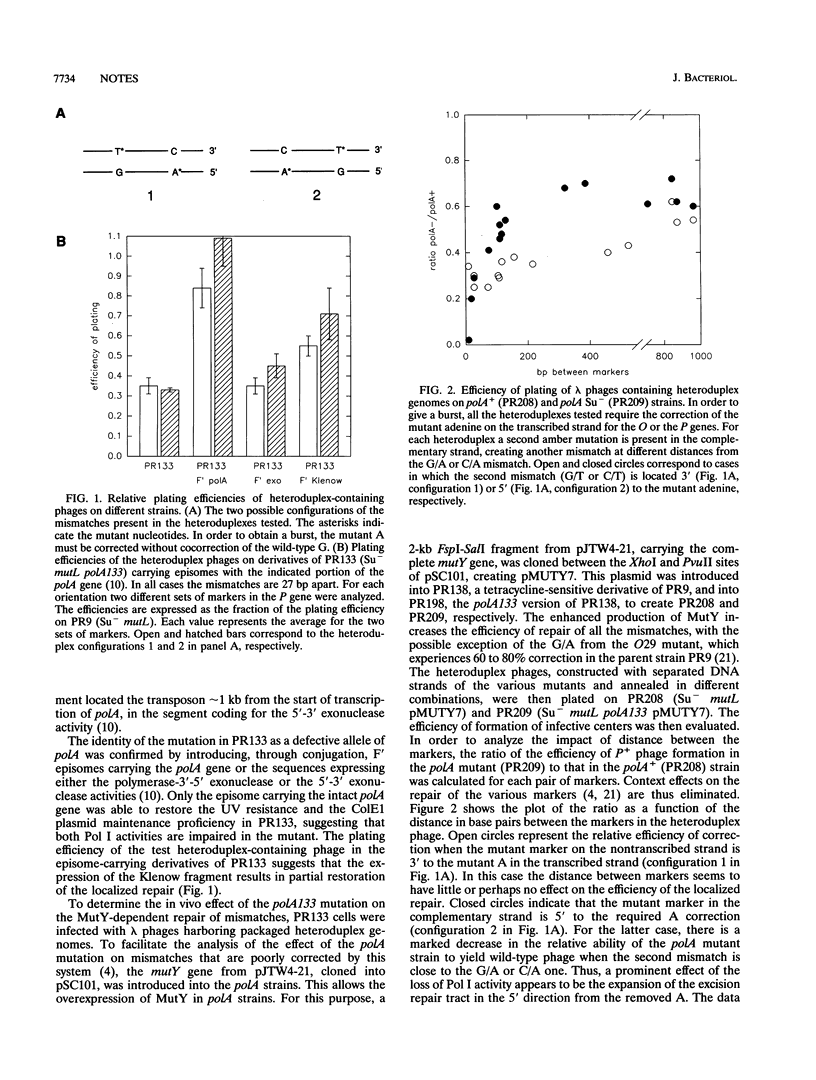
In vivo experiments with heteroduplex lambda genomes show that the MutY mismatch repair system of Escherichia coli defines an average repair tract that is shorter than 27 nucleotides and longer than 9 nucleotides and extends 3' from the corrected adenine. The phenotype of a mutant defective in DNA polymerase I shows that this enzyme plays a significant, though not an essential, role in the in vivo repair of apurinic sites generated by this system. Evidence is presented that in the absence of polymerase I the repair tracts are modestly longer than in the polA+ extending in the 5' direction from the corrected adenine, suggesting a role for another DNA polymerase.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Au K. G., Cabrera M., Miller J. H., Modrich P. Escherichia coli mutY gene product is required for specific A-G----C.G mismatch correction. Proc Natl Acad Sci U S A. 1988 Dec;85(23):9163–9166. doi: 10.1073/pnas.85.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au K. G., Clark S., Miller J. H., Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper P. K., Hanawalt P. C. Role of DNA polymerase I and the rec system in excision-repair in Escherichia coli. Proc Natl Acad Sci U S A. 1972 May;69(5):1156–1160. doi: 10.1073/pnas.69.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzidic S., Radman M. Genetic requirements for hyper-recombination by very short patch mismatch repair: involvement of Escherichia coli DNA polymerase I. Mol Gen Genet. 1989 Jun;217(2-3):254–256. doi: 10.1007/BF02464889. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Grindley N. D. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984 May;158(2):636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lieb M. Specific mismatch correction in bacteriophage lambda crosses by very short patch repair. Mol Gen Genet. 1983;191(1):118–125. doi: 10.1007/BF00330898. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Chang D. Y. A novel nucleotide excision repair for the conversion of an A/G mismatch to C/G base pair in E. coli. Cell. 1988 Sep 9;54(6):805–812. doi: 10.1016/s0092-8674(88)91109-9. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Chang D. Y. Repair of single base-pair transversion mismatches of Escherichia coli in vitro: correction of certain A/G mismatches is independent of dam methylation and host mutHLS gene functions. Genetics. 1988 Apr;118(4):593–600. doi: 10.1093/genetics/118.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Cuipa M. J., Ip M. S., Shanabruch W. G. Specific A/G-to-C.G mismatch repair in Salmonella typhimurium LT2 requires the mutB gene product. J Bacteriol. 1990 Mar;172(3):1232–1240. doi: 10.1128/jb.172.3.1232-1240.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Cruz C., Grollman A. P., Miller J. H. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels M. L., Pham L., Nghiem Y., Cruz C., Miller J. H. MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Res. 1990 Jul 11;18(13):3841–3845. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- Radicella J. P., Clark E. A., Fox M. S. Some mismatch repair activities in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9674–9678. doi: 10.1073/pnas.85.24.9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard M., Lefevre J. C., Mostachfi P., Gasc A. M., Sarda C. Localized conversion in Streptococcus pneumoniae recombination: heteroduplex preference. Genetics. 1985 Aug;110(4):557–568. doi: 10.1093/genetics/110.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. S., Lahue R. S., Au K. G., Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988 May 15;263(14):6829–6835. [PubMed] [Google Scholar]
- Tiraby J. G., Fox M. S. Marker discrimination in transformation and mutation of pneumococcus. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3541–3545. doi: 10.1073/pnas.70.12.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai-Wu J. J., Liu H. F., Lu A. L. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A.C and A.G mispairs. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8779–8783. doi: 10.1073/pnas.89.18.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Weinberger S., Sperling J. Characterization of Escherichia coli mutant strains deficient in AP DNA-repair synthesis. Mutat Res. 1986 Sep;166(2):123–134. doi: 10.1016/0167-8817(86)90010-6. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. In vitro correction of G.T mispairs to G.C pairs in nuclear extracts from human cells. Nature. 1989 May 18;339(6221):234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. Mismatch-specific thymine DNA glycosylase and DNA polymerase beta mediate the correction of G.T mispairs in nuclear extracts from human cells. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5842–5845. doi: 10.1073/pnas.87.15.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y. C., Chang D. Y., Masin J., Lu A. L. Two nicking enzyme systems specific for mismatch-containing DNA in nuclear extracts from human cells. J Biol Chem. 1991 Apr 5;266(10):6480–6484. [PubMed] [Google Scholar]


