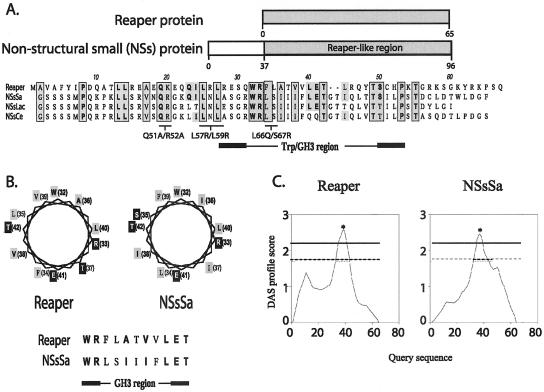
Figure 1.
Sequence alignment of NSs and Reaper. (a) Public databases queried for Reaper homologues using a tBLASTn algorithm yielded a group of nonstructural proteins encoded by the small genome segment (NSs) expressed in the Bunyaviridae viral family. The Reaper-like region in the NSs protein is highlighted in the schematic diagram of the NSs. Areas of similarity between the NSs and Reaper are shaded. Amino acid identities are represented in bold. Areas of similarity and identity between the three NSs proteins and Reaper are boxed. Also indicated are areas of sequence similarity mutated and assayed for function (see Figure 3). (b) Helical wheel projection diagram of the Reaper and NSsSa GH3 region, which is predicted to conform to an amphipathic α-helix. Similar and identical residues are represented in bold. Hydrophobic residues are shaded in gray, and hydrophilic residues in black. (c) Hydrophobicity analysis of Reaper and the NSsSa RLR by DAS TM-segment prediction. Dashed horizontal line correspond to loose cutoff DAS score of 1.7, and the solid gray line corresponds to the strict cutoff DAS score of 2.2. Starred area of high hydrophobicity corresponds to amino acids 33–42 (in both Reaper and NSs RLR).