Abstract
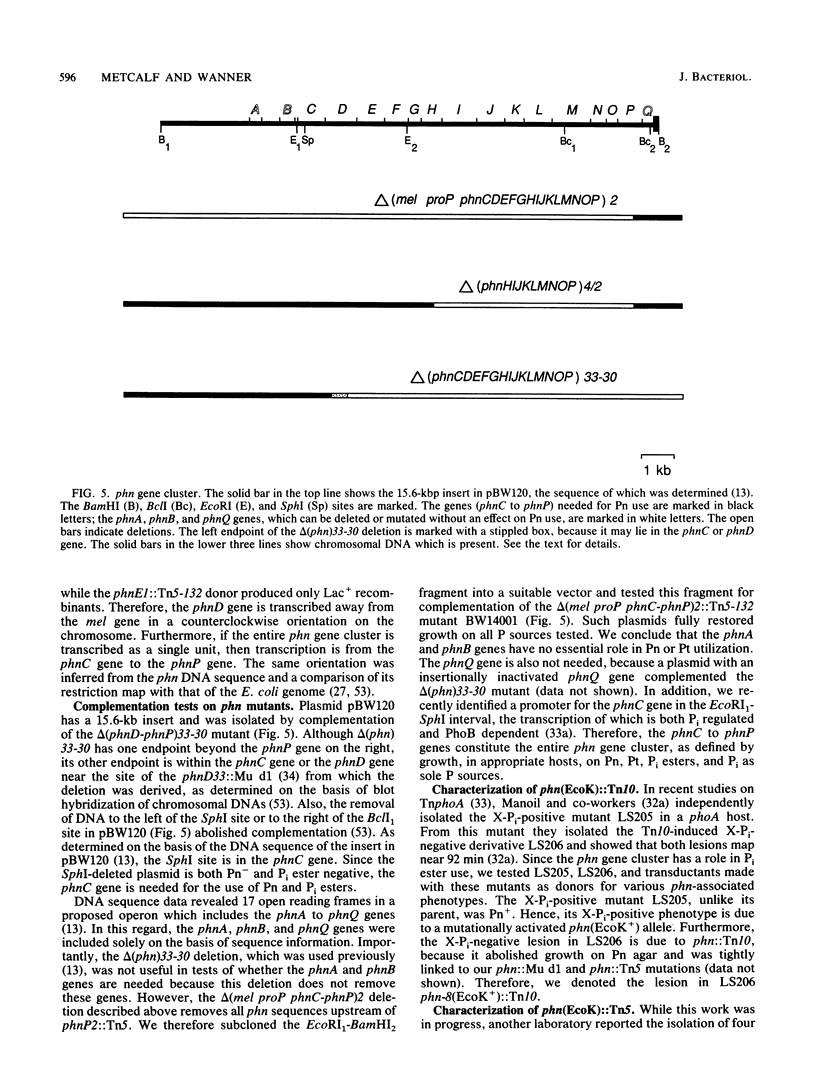
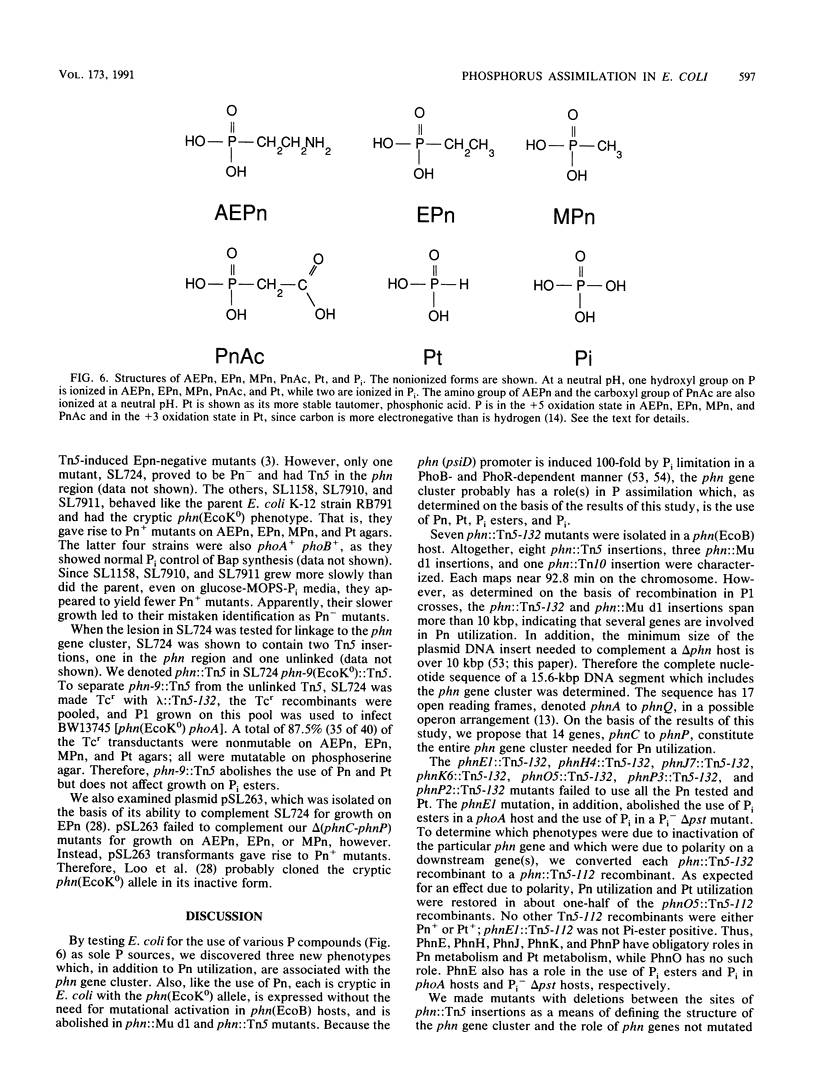
The phn (psiD) gene cluster is induced during Pi limitation and is required for the use of phosphonates (Pn) as a phosphorus (P) source. Twelve independent Pn-negative (Pn-) mutants have lesions in the phn gene cluster which, as determined on the basis of recombination frequencies, is larger than 10 kbp. This distance formed the basis for determining the complete DNA sequence of a 15.6-kbp BamHI fragment, the sequences of which suggested an operon with 17 open reading frames, denoted (in alphabetical order) the phnA to phnQ genes (C.-M. Chen, Q.-Z. Ye, Z. Zhu, B. L. Wanner, and C. T. Walsh, J. Biol. Chem. 265:4461-4471, 1990) Ten Pn- lesions lie in the phnD, phnE, phnH, phnJ, phnK, phnO, and phnP genes. We propose a smaller gene cluster with 14 open reading frames, phnC to phnP, which probably encode transporter and regulatory functions, in addition to proteins needed in Pn biodegradation. On the basis of the effects on phosphite (Pt), Pi ester, and Pi use, we propose that PhnC, PhnD, and PhnE constitute a binding protein-dependent Pn transporter which also transports Pt, Pi esters, and Pi. We propose that PhnO has a regulatory role because a phnO lesion affects no biochemical function, except for those due to polarity. Presumably, the 10 other phn gene products mostly act in an enzyme complex needed for breaking the stable carbon-phosphorus bond. Interestingly, all Pn- mutations abolish the use not only of Pn but also of Pt, in which P is in the +3 oxidation state. Therefore, Pn metabolism and Pt metabolism are related, supporting a biochemical mechanism for carbon-phosphorus bond cleavage which involves redox chemistry at the P center. Furthermore, our discovery of Pi-regulated genes for the assimilation of reduced P suggests that a P redox cycle may be important in biology.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agrawal D. K., Wanner B. L. A phoA structural gene mutation that conditionally affects formation of the enzyme bacterial alkaline phosphatase. J Bacteriol. 1990 Jun;172(6):3180–3190. doi: 10.1128/jb.172.6.3180-3190.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames G. F. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- Bell A. W., Buckel S. D., Groarke J. M., Hope J. N., Kingsley D. H., Hermodson M. A. The nucleotide sequences of the rbsD, rbsA, and rbsC genes of Escherichia coli K12. J Biol Chem. 1986 Jun 15;261(17):7652–7658. [PubMed] [Google Scholar]
- Benz R., Schmid A., Van der Ley P., Tommassen J. Molecular basis of porin selectivity: membrane experiments with OmpC-PhoE and OmpF-PhoE hybrid proteins of Escherichia coli K-12. Biochim Biophys Acta. 1989 May 19;981(1):8–14. doi: 10.1016/0005-2736(89)90075-8. [DOI] [PubMed] [Google Scholar]
- Berg D. E. Control of gene expression by a mobile recombinational switch. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4880–4884. doi: 10.1073/pnas.77.8.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzoska P., Boos W. Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the ugp transport system of Escherichia coli. J Bacteriol. 1988 Sep;170(9):4125–4135. doi: 10.1128/jb.170.9.4125-4135.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASIDA L. E., Jr Microbial oxidation and utilization of orthophosphite during growth. J Bacteriol. 1960 Aug;80:237–241. doi: 10.1128/jb.80.2.237-241.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellazzi M., Brachet P., Eisen H. Isolation and characterization of deletions in bacteriophage lambda residing as prophage in E. coli K 12. Mol Gen Genet. 1972;117(3):211–218. doi: 10.1007/BF00271648. [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. M., Ye Q. Z., Zhu Z. M., Wanner B. L., Walsh C. T. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J Biol Chem. 1990 Mar 15;265(8):4461–4471. [PubMed] [Google Scholar]
- Cox G. B., Webb D., Rosenberg H. Specific amino acid residues in both the PstB and PstC proteins are required for phosphate transport by the Escherichia coli Pst system. J Bacteriol. 1989 Mar;171(3):1531–1534. doi: 10.1128/jb.171.3.1531-1534.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos G. F., Walker G. C., Signer E. R. Genetic manipulations in Rhizobium meliloti utilizing two new transposon Tn5 derivatives. Mol Gen Genet. 1986 Sep;204(3):485–491. doi: 10.1007/BF00331029. [DOI] [PubMed] [Google Scholar]
- Ehrmann M., Boos W., Ormseth E., Schweizer H., Larson T. J. Divergent transcription of the sn-glycerol-3-phosphate active transport (glpT) and anaerobic sn-glycerol-3-phosphate dehydrogenase (glpA glpC glpB) genes of Escherichia coli K-12. J Bacteriol. 1987 Feb;169(2):526–532. doi: 10.1128/jb.169.2.526-532.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. L., Winans L., Jr, Helms S. J. Anaerobic utilization of phosphite and hypophosphite by Bacillus sp. Appl Environ Microbiol. 1978 May;35(5):937–944. doi: 10.1128/aem.35.5.937-944.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M. J., Kadner R. J. Nucleotide sequence of the uhp region of Escherichia coli. J Bacteriol. 1987 Aug;169(8):3556–3563. doi: 10.1128/jb.169.8.3556-3563.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnant M. K., Stauffer G. V. Construction and analysis of plasmids containing the Escherichia coli serB gene. Mol Gen Genet. 1984;193(1):72–75. doi: 10.1007/BF00327416. [DOI] [PubMed] [Google Scholar]
- Gierasch L. M. Signal sequences. Biochemistry. 1989 Feb 7;28(3):923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Gowrishankar J. Nucleotide sequence of the osmoregulatory proU operon of Escherichia coli. J Bacteriol. 1989 Apr;171(4):1923–1931. doi: 10.1128/jb.171.4.1923-1931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Loo S. H., Peters N. K., Frost J. W. Genetic characterization of an Escherichia coli mutant deficient in organophosphonate biodegradation. Biochem Biophys Res Commun. 1987 Oct 14;148(1):148–152. doi: 10.1016/0006-291x(87)91088-6. [DOI] [PubMed] [Google Scholar]
- Makino K., Shinagawa H., Amemura M., Kawamoto T., Yamada M., Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989 Dec 5;210(3):551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- Makino K., Shinagawa H., Amemura M., Nakata A. Nucleotide sequence of the phoB gene, the positive regulatory gene for the phosphate regulon of Escherichia coli K-12. J Mol Biol. 1986 Jul 5;190(1):37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- Malacinski G. M., Konetzka W. A. Orthophosphite-nicotinamide adenine dinucleotide oxidoreductase from Pseudomonas fluorescens. J Bacteriol. 1967 Jun;93(6):1906–1910. doi: 10.1128/jb.93.6.1906-1910.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malacinski G., Konetzka W. A. Bacterial oxidation of orthophosphate. J Bacteriol. 1966 Feb;91(2):578–582. doi: 10.1128/jb.91.2.578-582.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986 Sep 26;233(4771):1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- Metcalf W. W., Steed P. M., Wanner B. L. Identification of phosphate starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J Bacteriol. 1990 Jun;172(6):3191–3200. doi: 10.1128/jb.172.6.3191-3200.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K., Higaki N., Kimura A. A microbial carbon-phosphorus bond cleavage enzyme requires two protein components for activity. J Bacteriol. 1989 Aug;171(8):4504–4506. doi: 10.1128/jb.171.8.4504-4506.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., Huang H. V., Berg D. E. Bidirectional chain-termination nucleotide sequencing: transposon Tn5seq1 as a mobile source of primer sites. Gene. 1988 Apr 15;64(1):135–145. doi: 10.1016/0378-1119(88)90487-8. [DOI] [PubMed] [Google Scholar]
- Nohno T., Saito T., Hong J. S. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ). Mol Gen Genet. 1986 Nov;205(2):260–269. doi: 10.1007/BF00430437. [DOI] [PubMed] [Google Scholar]
- Olsen D. B., Hepburn T. W., Moos M., Mariano P. S., Dunaway-Mariano D. Investigation of the Bacillus cereus phosphonoacetaldehyde hydrolase. Evidence for a Schiff base mechanism and sequence analysis of an active-site peptide containing the catalytic lysine residue. Biochemistry. 1988 Mar 22;27(6):2229–2234. doi: 10.1021/bi00406a063. [DOI] [PubMed] [Google Scholar]
- Overduin P., Boos W., Tommassen J. Nucleotide sequence of the ugp genes of Escherichia coli K-12: homology to the maltose system. Mol Microbiol. 1988 Nov;2(6):767–775. doi: 10.1111/j.1365-2958.1988.tb00088.x. [DOI] [PubMed] [Google Scholar]
- Pecher A., Zinoni F., Jatisatienr C., Wirth R., Hennecke H., Böck A. On the redox control of synthesis of anaerobically induced enzymes in enterobacteriaceae. Arch Microbiol. 1983 Nov;136(2):131–136. doi: 10.1007/BF00404787. [DOI] [PubMed] [Google Scholar]
- Schowanek D., Verstraete W. Phosphonate utilization by bacterial cultures and enrichments from environmental samples. Appl Environ Microbiol. 1990 Apr;56(4):895–903. doi: 10.1128/aem.56.4.895-903.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scripture J. B., Voelker C., Miller S., O'Donnell R. T., Polgar L., Rade J., Horazdovsky B. F., Hogg R. W. High-affinity L-arabinose transport operon. Nucleotide sequence and analysis of gene products. J Mol Biol. 1987 Sep 5;197(1):37–46. doi: 10.1016/0022-2836(87)90607-3. [DOI] [PubMed] [Google Scholar]
- Wackett L. P., Shames S. L., Venditti C. P., Walsh C. T. Bacterial carbon-phosphorus lyase: products, rates, and regulation of phosphonic and phosphinic acid metabolism. J Bacteriol. 1987 Feb;169(2):710–717. doi: 10.1128/jb.169.2.710-717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett L. P., Wanner B. L., Venditti C. P., Walsh C. T. Involvement of the phosphate regulon and the psiD locus in carbon-phosphorus lyase activity of Escherichia coli K-12. J Bacteriol. 1987 Apr;169(4):1753–1756. doi: 10.1128/jb.169.4.1753-1756.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. Bacterial alkaline phosphatase clonal variation in some Escherichia coli K-12 phoR mutant strains. J Bacteriol. 1986 Dec;168(3):1366–1371. doi: 10.1128/jb.168.3.1366-1371.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L., Boline J. A. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J Bacteriol. 1990 Mar;172(3):1186–1196. doi: 10.1128/jb.172.3.1186-1196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. Control of phoR-dependent bacterial alkaline phosphatase clonal variation by the phoM region. J Bacteriol. 1987 Feb;169(2):900–903. doi: 10.1128/jb.169.2.900-903.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L., McSharry R. Phosphate-controlled gene expression in Escherichia coli K12 using Mudl-directed lacZ fusions. J Mol Biol. 1982 Jul 5;158(3):347–363. doi: 10.1016/0022-2836(82)90202-9. [DOI] [PubMed] [Google Scholar]
- Wanner B. L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986 Sep 5;191(1):39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- Wanner B. L. Overlapping and separate controls on the phosphate regulon in Escherichia coli K12. J Mol Biol. 1983 May 25;166(3):283–308. doi: 10.1016/s0022-2836(83)80086-2. [DOI] [PubMed] [Google Scholar]
- Wood J. M., Zadworny D. Characterization of an inducible porter required for L-proline catabolism by Escherichia coli K12. Can J Biochem. 1979 Oct;57(10):1191–1199. doi: 10.1139/o79-155. [DOI] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]


