Abstract
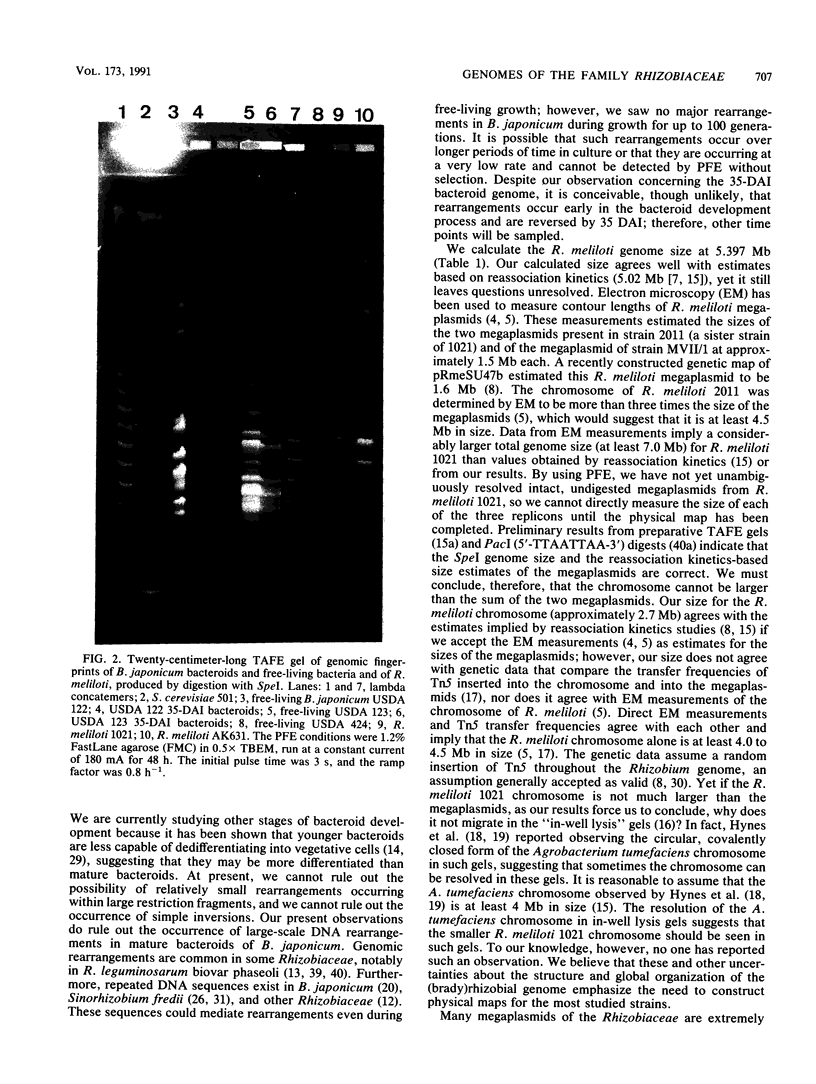
The lack of high-resolution genetic or physical maps for the family Rhizobiaceae limits our understanding of this agronomically important bacterial family. On the basis of statistical analyses of DNA sequences of the Rhizobiaceae and direct evaluation by pulsed-field agarose gel electrophoresis (PFE), five restriction endonucleases with AT-rich target sites were identified as the most rarely cutting: AseI (5'-ATTAAT-3'), DraI (5'-TTTAAA-3'), SpeI (5'-ACTAGT-3'), SspI (5'-AATAAT-3'), and XbaI (5'-TCTAGA-3'). We computed the sizes of the genomes of Bradyrhizobium japonicum USDA 424 and Rhizobium meliloti 1021 by adding the sizes of DNA fragments generated by SpeI digests. The genome sizes of R. meliloti 1021 and B. japonicum USDA 424 were 5,379 +/- 282.5 kb and 6,195 +/- 192.4 kb, respectively. We also compared the organization of the genomes of free-living and bacteroid forms of B. japonicum. No differences between the PFE-resolved genomic fingerprints of free-living and mature (35 days after inoculation) bacteroids of B. japonicum USDA 123 and USDA 122 were observed. Also, B. japonicum USDA 123 genomic fingerprints were unchanged after passage through nodules and after maintenance on a rich growth medium for 100 generations. We conclude that large-scale DNA rearrangements are not seen in mature bacteroids or during free-living growth on rich growth media under laboratory conditions.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almagor H. A Markov analysis of DNA sequences. J Theor Biol. 1983 Oct 21;104(4):633–645. doi: 10.1016/0022-5193(83)90251-5. [DOI] [PubMed] [Google Scholar]
- BERNS K. I., THOMAS C. A., Jr ISOLATION OF HIGH MOLECULAR WEIGHT DNA FROM HEMOPHILUS INFLUENZAE. J Mol Biol. 1965 Mar;11:476–490. doi: 10.1016/s0022-2836(65)80004-3. [DOI] [PubMed] [Google Scholar]
- Burkardt B., Burkardt H. J. Visualization and exact molecular weight determination of a Rhizobium meliloti megaplasmid. J Mol Biol. 1984 May 15;175(2):213–218. doi: 10.1016/0022-2836(84)90475-3. [DOI] [PubMed] [Google Scholar]
- Burkardt B., Schillik D., Pühler A. Physical characterization of Rhizobium meliloti megaplasmids. Plasmid. 1987 Jan;17(1):13–25. doi: 10.1016/0147-619x(87)90004-7. [DOI] [PubMed] [Google Scholar]
- Carlson T. A., Guerinot M. L., Chelm B. K. Characterization of the gene encoding glutamine synthetase I (glnA) from Bradyrhizobium japonicum. J Bacteriol. 1985 May;162(2):698–703. doi: 10.1128/jb.162.2.698-703.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles T. C., Finan T. M. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J Bacteriol. 1990 May;172(5):2469–2476. doi: 10.1128/jb.172.5.2469-2476.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard W. G. Why do bacterial plasmids carry some genes and not others? Plasmid. 1989 May;21(3):167–174. doi: 10.1016/0147-619x(89)90040-1. [DOI] [PubMed] [Google Scholar]
- Finan T. M., Kunkel B., De Vos G. F., Signer E. R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986 Jul;167(1):66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M., González V., Brom S., Martínez E., Piñero D., Romero D., Dávila G., Palacios R. Reiterated DNA sequences in Rhizobium and Agrobacterium spp. J Bacteriol. 1987 Dec;169(12):5782–5788. doi: 10.1128/jb.169.12.5782-5788.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M., González V., Pardo M. A., Leija A., Martínez E., Romero D., Piñero D., Dávila G., Palacios R. Genomic instability in Rhizobium phaseoli. J Bacteriol. 1988 Mar;170(3):1191–1196. doi: 10.1128/jb.170.3.1191-1196.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes M. F., Quandt J., O'Connell M. P., Pühler A. Direct selection for curing and deletion of Rhizobium plasmids using transposons carrying the Bacillus subtilis sacB gene. Gene. 1989 May 15;78(1):111–120. doi: 10.1016/0378-1119(89)90319-3. [DOI] [PubMed] [Google Scholar]
- Hynes M. F., Simon R., Pühler A. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtC58. Plasmid. 1985 Mar;13(2):99–105. doi: 10.1016/0147-619x(85)90062-9. [DOI] [PubMed] [Google Scholar]
- Kaluza K., Hahn M., Hennecke H. Repeated sequences similar to insertion elements clustered around the nif region of the Rhizobium japonicum genome. J Bacteriol. 1985 May;162(2):535–542. doi: 10.1128/jb.162.2.535-542.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. J., Smith H. O. Sizing of the Haemophilus influenzae Rd genome by pulsed-field agarose gel electrophoresis. J Bacteriol. 1988 Sep;170(9):4402–4405. doi: 10.1128/jb.170.9.4402-4405.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MACHATTIE L. A., BERNE K. I., THOMAS C. A., Jr ELECTRON MICROSCOPY OF DNA FROM HEMOPHILUS INFLUENZAE. J Mol Biol. 1965 Mar;11:648–649. doi: 10.1016/s0022-2836(65)80019-5. [DOI] [PubMed] [Google Scholar]
- Martin M. O., Long S. R. Generalized transduction in Rhizobium meliloti. J Bacteriol. 1984 Jul;159(1):125–129. doi: 10.1128/jb.159.1.125-129.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson R. V., Atherly A. G. The presence of repeated DNA sequences and a partial restriction map of the pSym of Rhizobium fredii USDA193. Plasmid. 1986 Jul;16(1):37–44. doi: 10.1016/0147-619x(86)90077-6. [DOI] [PubMed] [Google Scholar]
- Mathew M. K., Smith C. L., Cantor C. R. High-resolution separation and accurate size determination in pulsed-field gel electrophoresis of DNA. 1. DNA size standards and the effect of agarose and temperature. Biochemistry. 1988 Dec 27;27(26):9204–9210. doi: 10.1021/bi00426a019. [DOI] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R. K., Atherly A. G. Reiteration of genes involved in symbiotic nitrogen fixation by fast-growing Rhizobium japonicum. J Bacteriol. 1984 Nov;160(2):785–787. doi: 10.1128/jb.160.2.785-787.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowsky M. J., Tully R. E., Cregan P. B., Keyser H. H. Genetic Diversity in Bradyrhizobium japonicum Serogroup 123 and Its Relation to Genotype-Specific Nodulation of Soybean. Appl Environ Microbiol. 1987 Nov;53(11):2624–2630. doi: 10.1128/aem.53.11.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Zidwick M. J., Abebe H. M. Bradyrhizobium japonicum Serocluster 123 and Diversity among Member Isolates. Appl Environ Microbiol. 1986 Jun;51(6):1212–1215. doi: 10.1128/aem.51.6.1212-1215.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Soberón-Chávez G., Nájera R., Olivera H., Segovia L. Genetic rearrangements of a Rhizobium phaseoli symbiotic plasmid. J Bacteriol. 1986 Aug;167(2):487–491. doi: 10.1128/jb.167.2.487-491.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobral B. W., Atherly A. G. A rapid and cost-effective method for preparing genomic DNA from gram-negative bacteria in agarose plugs for pulsed-field gel electrophoresis. Biotechniques. 1989 Oct;7(9):938–938. [PubMed] [Google Scholar]
- Sobral B. W., Sadowsky M. J., Atherly A. G. Genome analysis of Bradyrhizobium japonicum serocluster 123 field isolates by using field inversion gel electrophoresis. Appl Environ Microbiol. 1990 Jun;56(6):1949–1953. doi: 10.1128/aem.56.6.1949-1953.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanto A., Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1 genome: presence of two unique circular chromosomes. J Bacteriol. 1989 Nov;171(11):5850–5859. doi: 10.1128/jb.171.11.5850-5859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierny C., Iaccarino M. Comparative study of the symbiotic plasmid DNA in free living bacteria and bacteroids of Rhizobium leguminosarum. FEMS Microbiol Lett. 1989 Jul 1;51(1):15–19. doi: 10.1016/0378-1097(89)90069-4. [DOI] [PubMed] [Google Scholar]
- Zoon K. C., Scocca J. J. Constitution of the cell envelope of Haemophilus influenzae in relation to competence for genetic transformation. J Bacteriol. 1975 Aug;123(2):666–677. doi: 10.1128/jb.123.2.666-677.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]