Abstract
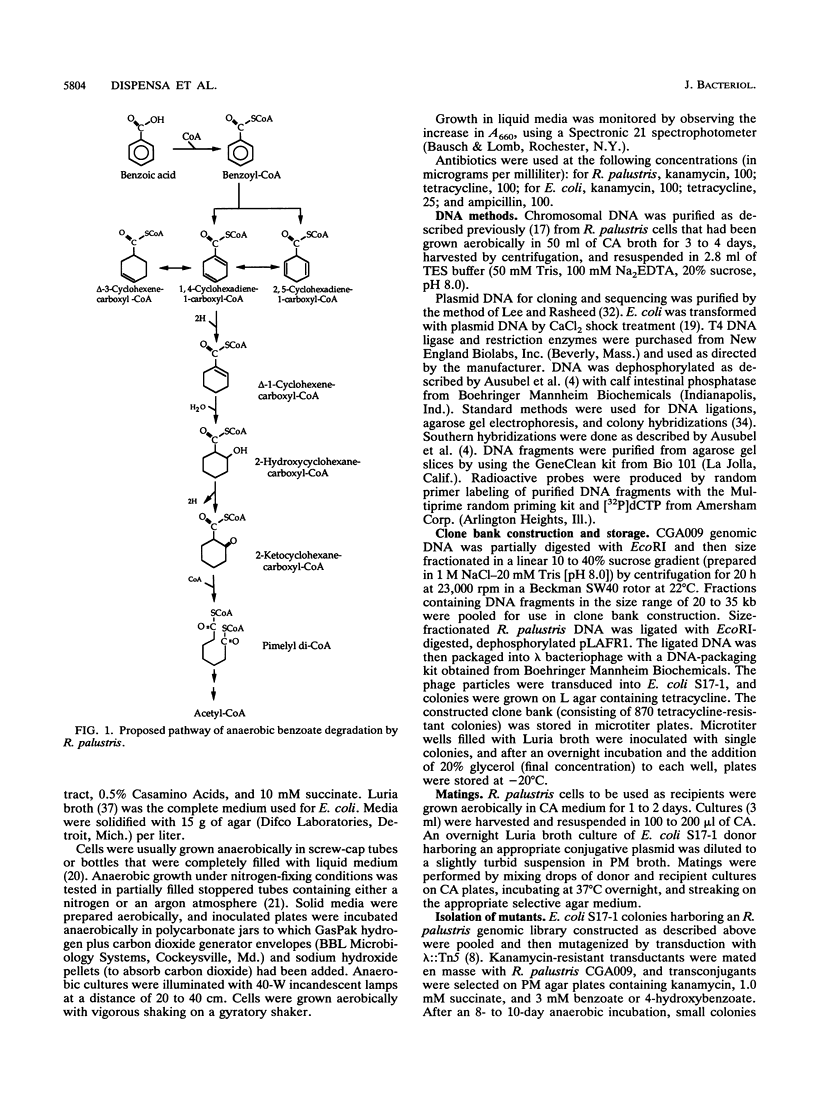
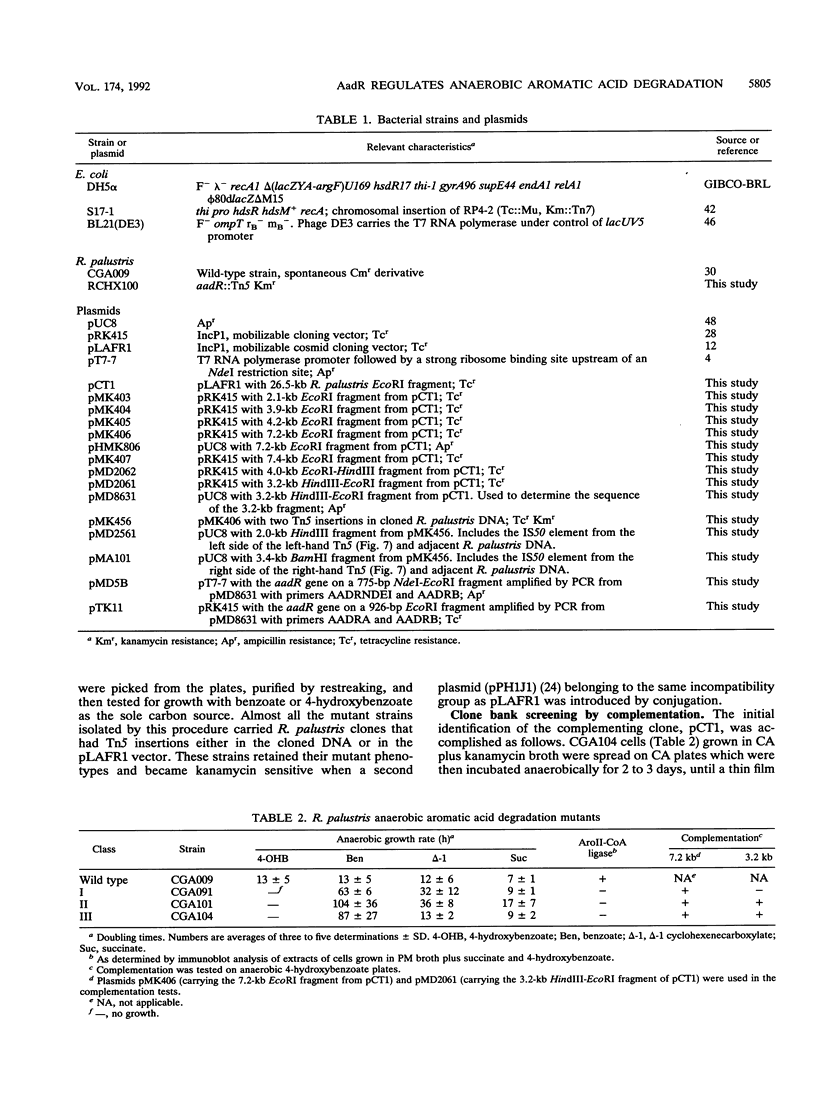
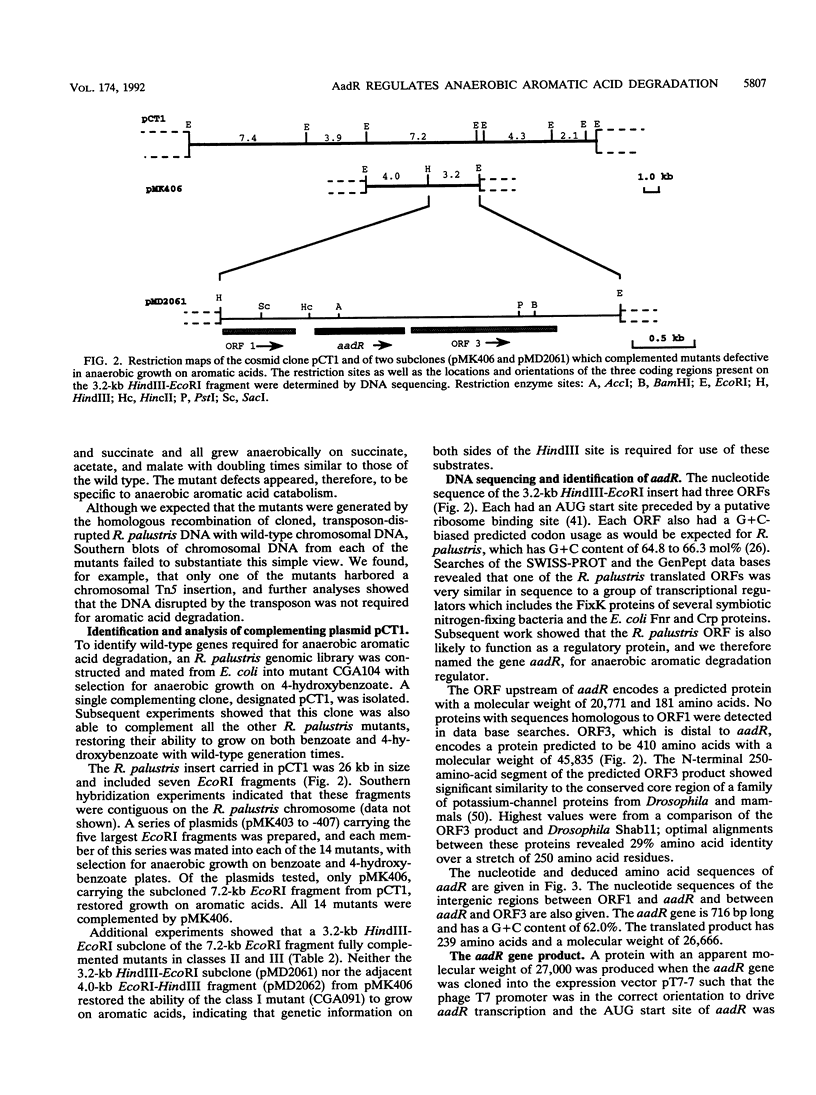
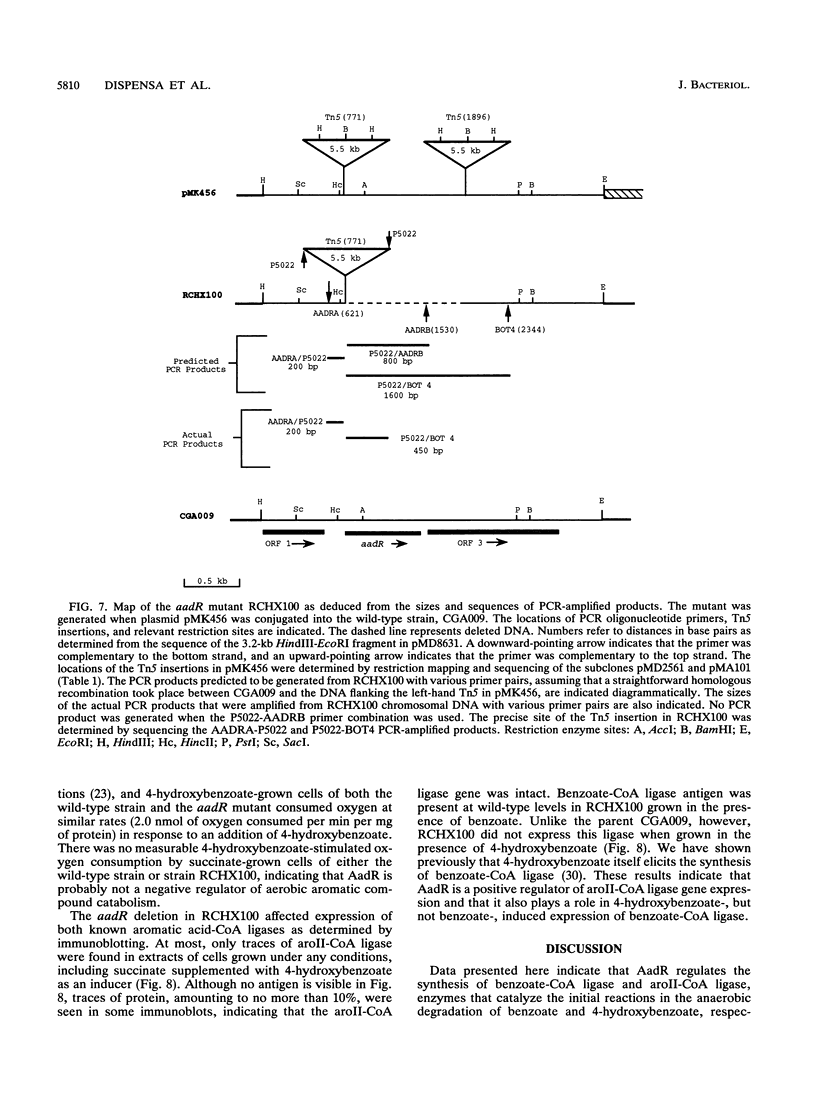
The purple nonsulfur phototrophic bacterium Rhodopseudomonas palustris converts structurally diverse aromatic carboxylic acids, including lignin monomers, to benzoate and 4-hydroxybenzoate under anaerobic conditions. These compounds are then further degraded via aromatic ring-fission pathways. A gene termed aadR, for anaerobic aromatic degradation regulator, was identified by complementation of mutants unable to grow anaerobically on 4-hydroxybenzoate. The deduced amino acid sequence of the aadR product is similar to a family of transcriptional regulators which includes Escherichia coli Fnr and Crp, Pseudomonas aeruginosa Anr, and rhizobial FixK and FixK-like proteins. A mutant with a deletion in aadR failed to grow on 4-hydroxybenzoate under anaerobic conditions and grew very slowly on benzoate. It also did not express aromatic acid-coenzyme A ligase II, an enzyme that catalyzes the first step of 4-hydroxybenzoate degradation, and it was defective in 4-hydroxybenzoate-induced expression of benzoate-coenzyme A ligase. The aadR deletion mutant was unaffected in other aspects of anaerobic growth. It grew normally on nonaromatic carbon sources and also under nitrogen-fixing conditions. In addition, aerobic growth on 4-hydroxybenzoate was indistinguishable from that of the wild type. These results indicate that AadR functions as a transcriptional activator of anaerobic aromatic acid degradation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenschmidt U., Oswald B., Fuchs G. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligases from a denitrifying Pseudomonas sp. J Bacteriol. 1991 Sep;173(17):5494–5501. doi: 10.1128/jb.173.17.5494-5501.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthamatten D., Scherb B., Hennecke H. Characterization of a fixLJ-regulated Bradyrhizobium japonicum gene sharing similarity with the Escherichia coli fnr and Rhizobium meliloti fixK genes. J Bacteriol. 1992 Apr;174(7):2111–2120. doi: 10.1128/jb.174.7.2111-2120.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerswald E. A., Ludwig G., Schaller H. Structural analysis of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):107–113. doi: 10.1101/sqb.1981.045.01.019. [DOI] [PubMed] [Google Scholar]
- Batut J., Daveran-Mingot M. L., David M., Jacobs J., Garnerone A. M., Kahn D. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 1989 Apr;8(4):1279–1286. doi: 10.1002/j.1460-2075.1989.tb03502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna-Romano S., Arnold W., Schlüter A., Boistard P., Pühler A., Priefer U. B. An Fnr-like protein encoded in Rhizobium leguminosarum biovar viciae shows structural and functional homology to Rhizobium meliloti FixK. Mol Gen Genet. 1990 Aug;223(1):138–147. doi: 10.1007/BF00315806. [DOI] [PubMed] [Google Scholar]
- Dutton P. L., Evans W. C. The metabolism of aromatic compounds by Rhodopseudomonas palustris. A new, reductive, method of aromatic ring metabolism. Biochem J. 1969 Jul;113(3):525–536. doi: 10.1042/bj1130525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans W. C., Fuchs G. Anaerobic degradation of aromatic compounds. Annu Rev Microbiol. 1988;42:289–317. doi: 10.1146/annurev.mi.42.100188.001445. [DOI] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Geissler J. F., Harwood C. S., Gibson J. Purification and properties of benzoate-coenzyme A ligase, a Rhodopseudomonas palustris enzyme involved in the anaerobic degradation of benzoate. J Bacteriol. 1988 Apr;170(4):1709–1714. doi: 10.1128/jb.170.4.1709-1714.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson K. J., Gibson J. Potential early intermediates in anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl Environ Microbiol. 1992 Feb;58(2):696–698. doi: 10.1128/aem.58.2.696-698.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckler R., Tschech A., Fuchs G. Reductive dehydroxylation of 4-hydroxybenzoyl-CoA to benzoyl-CoA in a denitrifying, phenol-degrading Pseudomonas species. FEBS Lett. 1989 Jul 17;251(1-2):237–240. doi: 10.1016/0014-5793(89)81461-9. [DOI] [PubMed] [Google Scholar]
- Gray K. M., Greenberg E. P. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J Bacteriol. 1992 Jul;174(13):4384–4390. doi: 10.1128/jb.174.13.4384-4390.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J., Trageser M., Six S., Unden G., Guest J. R. Characterization of the FNR protein of Escherichia coli, an iron-binding transcriptional regulator. Proc Biol Sci. 1991 May 22;244(1310):137–144. doi: 10.1098/rspb.1991.0062. [DOI] [PubMed] [Google Scholar]
- Harwood C. S., Gibson J. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol. 1988 Mar;54(3):712–717. doi: 10.1128/aem.54.3.712-717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. S., Gibson J. Uptake of benzoate by Rhodopseudomonas palustris grown anaerobically in light. J Bacteriol. 1986 Feb;165(2):504–509. doi: 10.1128/jb.165.2.504-509.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hegeman G. D. The metabolism of p-hydroxybenzoate by Rhodopseudomonas palustris and its regulation. Arch Mikrobiol. 1967;59(1):143–148. doi: 10.1007/BF00406325. [DOI] [PubMed] [Google Scholar]
- Hirsch P. R., Beringer J. E. A physical map of pPH1JI and pJB4JI. Plasmid. 1984 Sep;12(2):139–141. doi: 10.1016/0147-619x(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Kaminski P. A., Mandon K., Arigoni F., Desnoues N., Elmerich C. Regulation of nitrogen fixation in Azorhizobium caulinodans: identification of a fixK-like gene, a positive regulator of nifA. Mol Microbiol. 1991 Aug;5(8):1983–1991. doi: 10.1111/j.1365-2958.1991.tb00820.x. [DOI] [PubMed] [Google Scholar]
- Keen N. T., Tamaki S., Kobayashi D., Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988 Oct 15;70(1):191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- Kiley P. J., Reznikoff W. S. Fnr mutants that activate gene expression in the presence of oxygen. J Bacteriol. 1991 Jan;173(1):16–22. doi: 10.1128/jb.173.1.16-22.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J., Fuchs G. Enzymatic reduction of benzoyl-CoA to alicyclic compounds, a key reaction in anaerobic aromatic metabolism. Eur J Biochem. 1992 Apr 1;205(1):195–202. doi: 10.1111/j.1432-1033.1992.tb16768.x. [DOI] [PubMed] [Google Scholar]
- Lee S. Y., Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. Biotechniques. 1990 Dec;9(6):676–679. [PubMed] [Google Scholar]
- MacInnes J. I., Kim J. E., Lian C. J., Soltes G. A. Actinobacillus pleuropneumoniae hlyX gene homology with the fnr gene of Escherichia coli. J Bacteriol. 1990 Aug;172(8):4587–4592. doi: 10.1128/jb.172.8.4587-4592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville S. B., Gunsalus R. P. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990 Nov 5;265(31):18733–18736. [PubMed] [Google Scholar]
- Merkel S. M., Eberhard A. E., Gibson J., Harwood C. S. Involvement of coenzyme A thioesters in anaerobic metabolism of 4-hydroxybenzoate by Rhodopseudomonas palustris. J Bacteriol. 1989 Jan;171(1):1–7. doi: 10.1128/jb.171.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. G. Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol Microbiol. 1991 Jun;5(6):1469–1481. doi: 10.1111/j.1365-2958.1991.tb00793.x. [DOI] [PubMed] [Google Scholar]
- Sharrocks A. D., Green J., Guest J. R. In vivo and in vitro mutants of FNR the anaerobic transcriptional regulator of E. coli. FEBS Lett. 1990 Sep 17;270(1-2):119–122. doi: 10.1016/0014-5793(90)81248-m. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Spiro S., Gaston K. L., Bell A. I., Roberts R. E., Busby S. J., Guest J. R. Interconversion of the DNA-binding specificities of two related transcription regulators, CRP and FNR. Mol Microbiol. 1990 Nov;4(11):1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev. 1990 Aug;6(4):399–428. doi: 10.1111/j.1574-6968.1990.tb04109.x. [DOI] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Trageser M., Unden G. Role of cysteine residues and of metal ions in the regulatory functioning of FNR, the transcriptional regulator of anaerobic respiration in Escherichia coli. Mol Microbiol. 1989 May;3(5):593–599. doi: 10.1111/j.1365-2958.1989.tb00206.x. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987 Nov 20;198(2):311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- Wei A., Covarrubias M., Butler A., Baker K., Pak M., Salkoff L. K+ current diversity is produced by an extended gene family conserved in Drosophila and mouse. Science. 1990 May 4;248(4955):599–603. doi: 10.1126/science.2333511. [DOI] [PubMed] [Google Scholar]
- Zimmermann A., Reimmann C., Galimand M., Haas D. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol Microbiol. 1991 Jun;5(6):1483–1490. doi: 10.1111/j.1365-2958.1991.tb00794.x. [DOI] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]