Abstract
Uracil-DNA glycosylase activity was found in Streptococcus pneumoniae, and the enzyme was partially purified. An ung mutant lacking the activity was obtained by positive selection of cells transformed with a plasmid containing uracil in its DNA. The effects of the ung mutation on mutagenic processes in S. pneumoniae were examined. The sequence of several malM mutations revertible by nitrous acid showed them to correspond to A.T----G.C transitions. This confirmed a prior deduction that nitrous acid action on transforming DNA gave only G.C----A.T mutations. Examination of malM mutant reversion frequencies in ung strains indicated that G.C----A.T mutation rates generally were 10-fold higher than in wild-type strains, presumably owing to lack of repair of deaminated cytosine residues in DNA. No effect of ung on mutation avoidance by the Hex mismatch repair system was observed, which means that uracil incorporation and removal from nascent DNA cannot be solely responsible for producing strand breaks that target nascent DNA for correction after replication. One malM mutation corresponding to an A.T----G.C transition showed a 10-fold-higher spontaneous reversion frequency than other such transitions in a wild-type background. This "hot spot" was located in a directly repeated DNA sequence; it is proposed that transient slippage to the wild-type repeat during replication accounts for the higher reversion frequency.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- BERNS K. I., THOMAS C. A., Jr ISOLATION OF HIGH MOLECULAR WEIGHT DNA FROM HEMOPHILUS INFLUENZAE. J Mol Biol. 1965 Mar;11:476–490. doi: 10.1016/s0022-2836(65)80004-3. [DOI] [PubMed] [Google Scholar]
- Ballester S., Lopez P., Alonso J. C., Espinosa M., Lacks S. A. Selective advantage of deletions enhancing chloramphenicol acetyltransferase gene expression in Streptococcus pneumoniae plasmids. Gene. 1986;41(2-3):153–163. doi: 10.1016/0378-1119(86)90094-6. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Burgers P. M., Klein M. B. Selection by genetic transformation of a Saccharomyces cerevisiae mutant defective for the nuclear uracil-DNA-glycosylase. J Bacteriol. 1986 Jun;166(3):905–913. doi: 10.1128/jb.166.3.905-913.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Roger M., Sicard A. M. Excision and repair of mismatched base pairs in transformation of Streptococcus pneumoniae. Mol Gen Genet. 1980 Apr;178(1):191–201. doi: 10.1007/BF00267229. [DOI] [PubMed] [Google Scholar]
- Currier T. C., Nester E. W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976 Dec;76(2):431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Chambers J. A. The cloning and overproduction of Escherichia coli uracil-DNA glycosylase. Gene. 1984 May;28(2):211–219. doi: 10.1016/0378-1119(84)90258-0. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Miller J. H. Mutagenic deamination of cytosine residues in DNA. Nature. 1980 Oct 9;287(5782):560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- Duncan B. K., Weiss B. Specific mutator effects of ung (uracil-DNA glycosylase) mutations in Escherichia coli. J Bacteriol. 1982 Aug;151(2):750–755. doi: 10.1128/jb.151.2.750-755.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasc A. M., Sicard A. M., Claverys J. P. Repair of single- and multiple-substitution mismatches during recombination in Streptococcus pneumoniae. Genetics. 1989 Jan;121(1):29–36. doi: 10.1093/genetics/121.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber L. T., Pang P. P., Sobell D. I., Mankovich J. A., Walker G. C. Nucleotide sequence of the Salmonella typhimurium mutS gene required for mismatch repair: homology of MutS and HexA of Streptococcus pneumoniae. J Bacteriol. 1988 Jan;170(1):197–202. doi: 10.1128/jb.170.1.197-202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T. Enzymatic excision of free hypoxanthine from polydeoxynucleotides and DNA containing deoxyinosine monophosphate residues. J Biol Chem. 1978 Sep 10;253(17):5877–5879. [PubMed] [Google Scholar]
- Kramer W., Kramer B., Williamson M. S., Fogel S. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J Bacteriol. 1989 Oct;171(10):5339–5346. doi: 10.1128/jb.171.10.5339-5346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
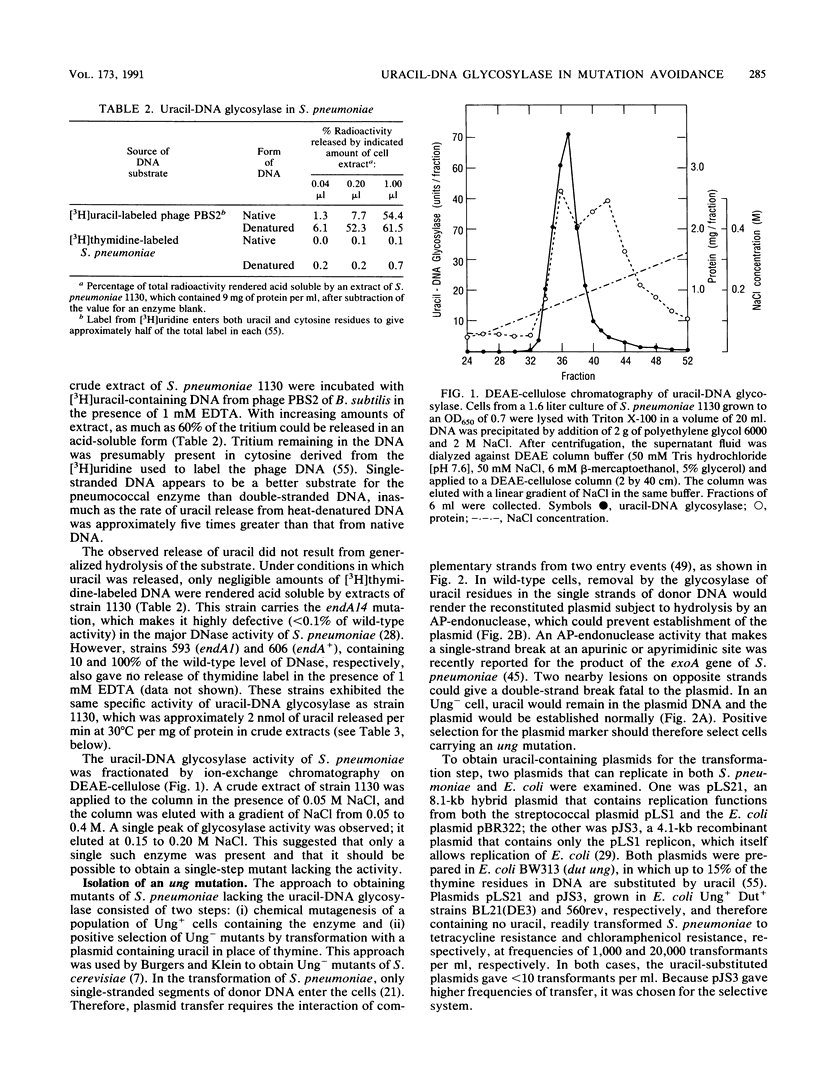
- LACKS S. Molecular fate of DNA in genetic transformation of Pneumococcus. J Mol Biol. 1962 Jul;5:119–131. doi: 10.1016/s0022-2836(62)80067-9. [DOI] [PubMed] [Google Scholar]
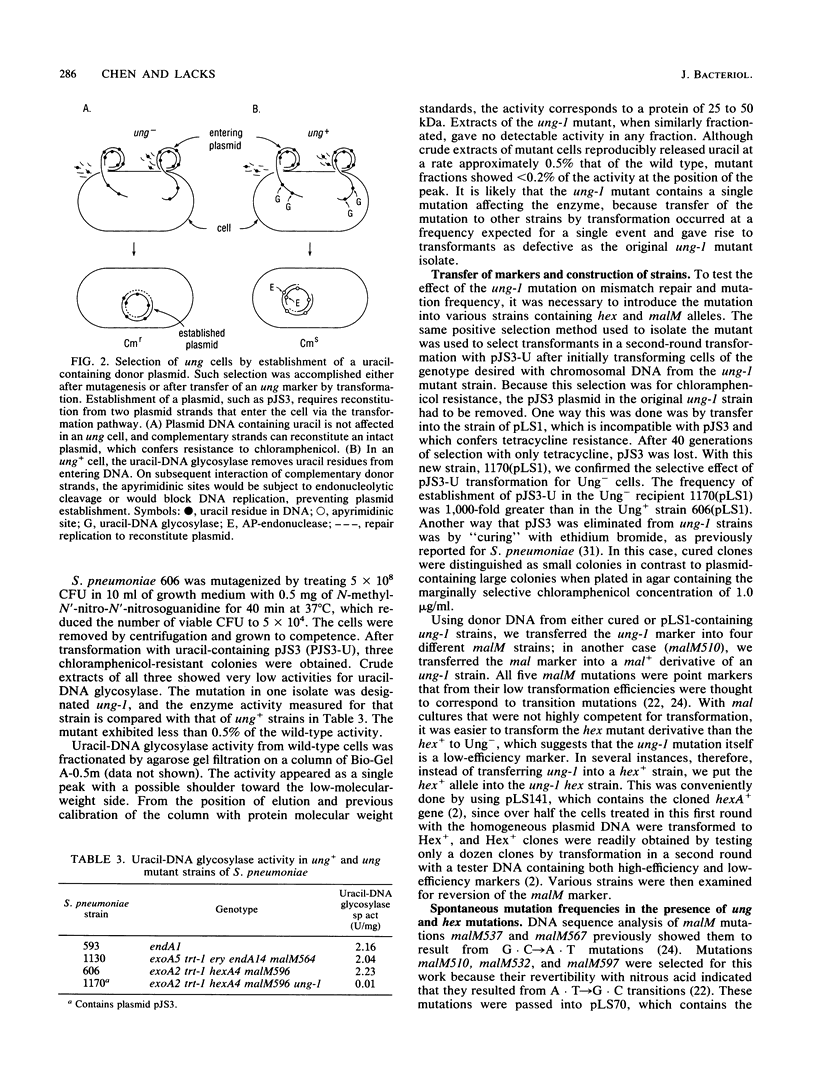
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lacks S. A., Dunn J. J., Greenberg B. Identification of base mismatches recognized by the heteroduplex-DNA-repair system of Streptococcus pneumoniae. Cell. 1982 Dec;31(2 Pt 1):327–336. doi: 10.1016/0092-8674(82)90126-x. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Lopez P., Greenberg B., Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986 Dec 20;192(4):753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Springhorn S. S. Cloning in Streptococcus pneumoniae of the gene for DpnII DNA methylase. J Bacteriol. 1984 Mar;157(3):934–936. doi: 10.1128/jb.157.3.934-936.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. A., Springhorn S. S. Transfer of recombinant plasmids containing the gene for DpnII DNA methylase into strains of Streptococcus pneumoniae that produce DpnI or DpnII restriction endonucleases. J Bacteriol. 1984 Jun;158(3):905–909. doi: 10.1128/jb.158.3.905-909.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J Biol Chem. 1975 Jun 10;250(11):4060–4066. [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Competence for deoxyribonucleic acid uptake and deoxyribonuclease action external to cells in the genetic transformation of Diplococcus pneumoniae. J Bacteriol. 1973 Apr;114(1):152–163. doi: 10.1128/jb.114.1.152-163.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B., Neuberger M. Identification of a deoxyribonuclease implicated in genetic transformation of Diplococcus pneumoniae. J Bacteriol. 1975 Jul;123(1):222–232. doi: 10.1128/jb.123.1.222-232.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Single-strand breakage on binding of DNA to cells in the genetic transformation of Diplococcus pneumoniae. J Mol Biol. 1976 Feb 25;101(2):255–275. doi: 10.1016/0022-2836(76)90376-4. [DOI] [PubMed] [Google Scholar]
- Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966 Jan;53(1):207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue R. S., Au K. G., Modrich P. DNA mismatch correction in a defined system. Science. 1989 Jul 14;245(4914):160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Lopez P., Espinosa M., Greenberg B., Lacks S. A. Generation of deletions in pneumococcal mal genes cloned in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5189–5193. doi: 10.1073/pnas.81.16.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P., Espinosa M., Greenberg B., Lacks S. A. Sulfonamide resistance in Streptococcus pneumoniae: DNA sequence of the gene encoding dihydropteroate synthase and characterization of the enzyme. J Bacteriol. 1987 Sep;169(9):4320–4326. doi: 10.1128/jb.169.9.4320-4326.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P., Espinosa M., Stassi D. L., Lacks S. A. Facilitation of plasmid transfer in Streptococcus pneumoniae by chromosomal homology. J Bacteriol. 1982 May;150(2):692–701. doi: 10.1128/jb.150.2.692-701.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankovich J. A., McIntyre C. A., Walker G. C. Nucleotide sequence of the Salmonella typhimurium mutL gene required for mismatch repair: homology of MutL to HexB of Streptococcus pneumoniae and to PMS1 of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989 Oct;171(10):5325–5331. doi: 10.1128/jb.171.10.5325-5331.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen L. C., Aasland R., Wittwer C. U., Krokan H. E., Helland D. E. Molecular cloning of human uracil-DNA glycosylase, a highly conserved DNA repair enzyme. EMBO J. 1989 Oct;8(10):3121–3125. doi: 10.1002/j.1460-2075.1989.tb08464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival K. J., Klein M. B., Burgers P. M. Molecular cloning and primary structure of the uracil-DNA-glycosylase gene from Saccharomyces cerevisiae. J Biol Chem. 1989 Feb 15;264(5):2593–2598. [PubMed] [Google Scholar]
- Priebe S. D., Hadi S. M., Greenberg B., Lacks S. A. Nucleotide sequence of the hexA gene for DNA mismatch repair in Streptococcus pneumoniae and homology of hexA to mutS of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1988 Jan;170(1):190–196. doi: 10.1128/jb.170.1.190-196.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhomme M., Martin B., Mejean V., Claverys J. P. Nucleotide sequence of the Streptococcus pneumoniae hexB mismatch repair gene: homology of HexB to MutL of Salmonella typhimurium and to PMS1 of Saccharomyces cerevisiae. J Bacteriol. 1989 Oct;171(10):5332–5338. doi: 10.1128/jb.171.10.5332-5338.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyet A., Greenberg B., Lacks S. A. Genetic and structural characterization of endA. A membrane-bound nuclease required for transformation of Streptococcus pneumoniae. J Mol Biol. 1990 Jun 20;213(4):727–738. doi: 10.1016/S0022-2836(05)80259-1. [DOI] [PubMed] [Google Scholar]
- Puyet A., Greenberg B., Lacks S. A. The exoA gene of Streptococcus pneumoniae and its product, a DNA exonuclease with apurinic endonuclease activity. J Bacteriol. 1989 May;171(5):2278–2286. doi: 10.1128/jb.171.5.2278-2286.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROMIG W. R. Infection of Bacillus subtilis with phenol-extracted bacteriophages. Virology. 1962 Apr;16:452–459. doi: 10.1016/0042-6822(62)90226-x. [DOI] [PubMed] [Google Scholar]
- Raimondo L. M., Lundh N. P., Martinez R. J. Primary adsorption site of phage PBS1: the flagellum of Bacillus subtilis. J Virol. 1968 Mar;2(3):256–264. doi: 10.1128/jvi.2.3.256-264.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C. W., Guild W. R. Monomer plasmid DNA transforms Streptococcus pneumoniae. Mol Gen Genet. 1981;181(1):57–62. doi: 10.1007/BF00339005. [DOI] [PubMed] [Google Scholar]
- Stassi D. L., Lopez P., Espinosa M., Lacks S. A. Cloning of chromosomal genes in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7028–7032. doi: 10.1073/pnas.78.11.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternglanz R., Wang H. F., Donegan J. J. Evidence that both growing DNA chains at a replication fork are synthesized discontinuously. Biochemistry. 1976 May 4;15(9):1838–1843. doi: 10.1021/bi00654a008. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tye B. K., Chien J., Lehman I. R., Duncan B. K., Warner H. R. Uracil incorporation: a source of pulse-labeled DNA fragments in the replication of the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1978 Jan;75(1):233–237. doi: 10.1073/pnas.75.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Hutcheon T., van de Sande J. H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J Biol Chem. 1988 Jun 5;263(16):7776–7784. [PubMed] [Google Scholar]
- Warner H. R., Duncan B. K., Garrett C., Neuhard J. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1981 Feb;145(2):687–695. doi: 10.1128/jb.145.2.687-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]


