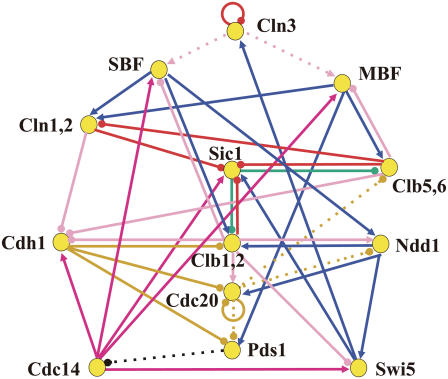
FIGURE 1.
Model of the reaction network that sustains the cell cycle of budding yeast. Each node represents a gene and its product, mRNA and protein. Arrows with a triangular head denote positive regulation, whereas arrows with a round head show negative regulation. Colors of arrows specify the types of regulation: transcriptional regulation (blue), phosphorylation (pink), dephosphorylation (dark pink), ubiquitination (yellow), phosphorylation as a mark of ubiquitination (red), protein-complex formation (green), and suppression of diffusion (black). Cdc28, which is CDK in budding yeast, is abundant through the cell cycle and hence is not explicitly considered in the model. Cln1 and Cln2 are assumed to work in combination and hence are treated as a unit (Cln1,2) in the model. Clb1,2 and Clb5,6 are also treated as units. The reactions indicated by dotted arrows are assumed to work only in specific stages: phosphorylation of SBF and MBF by Cln3 (stage 1), ubiquitination of Clb5,6, Ndd1, and Pds1 triggered by Cdc20 (stages 3–5), and suppression of diffusion of Cdc14 by Pds1 (stage 4).