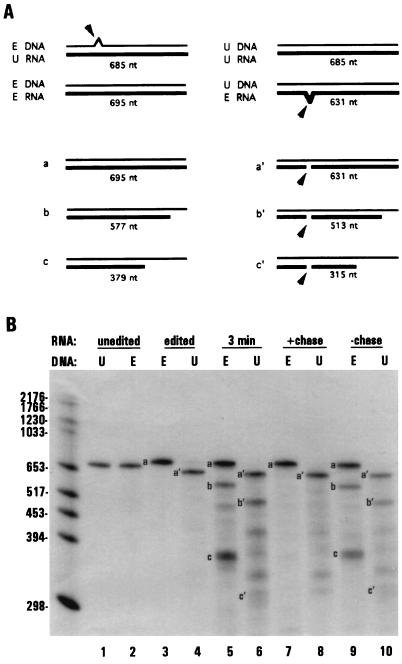
Figure 2.
RNAs associated with stalled transcription complexes are edited by dinucleotide insertion. (A) Schematic diagram of dinucleotide insertion assay. Arrows indicate dinucleotide bulges present in hybrids between edited (E) and unedited (U) molecules that are preferentially cleaved by S1 nuclease. Single nucleotide bulges are not cleaved efficiently and are therefore not depicted. Also shown are schematic diagrams of the protection patterns and observed sizes of labeled mitochondrial RNAs produced under limiting GTP conditions (thick lines). (B) Unlabeled ssDNA having either edited (E, probe 1, lanes 2, 3, 5, 7, and 9) or the corresponding unedited (U, probe 2, lanes 1, 4, 6, 8, and 10) coI sequence were used in S1 nuclease protection of labeled RNAs. Lanes 1–4, S1 protection of unedited (lanes 1 and 2) and edited (lanes 3 and 4) coI control transcripts. Lanes 5–10, RNAs synthesized in isolated mitochondria. Lanes 5 and 6, 3-min pulse labeling, limiting GTP; lanes 7 and 8, 3-min pulse labeling, limiting GTP, followed by a 12 min chase with 100 μM GTP; lanes 9 and 10, RNAs synthesized during a 15-min pulse labeling, limiting GTP. S1 protected samples were separated on a 4% denaturing polyacrylamide gel. The 5′ cleavage product resulting from S1 protection with probe 2 (U) was run off of the gel.