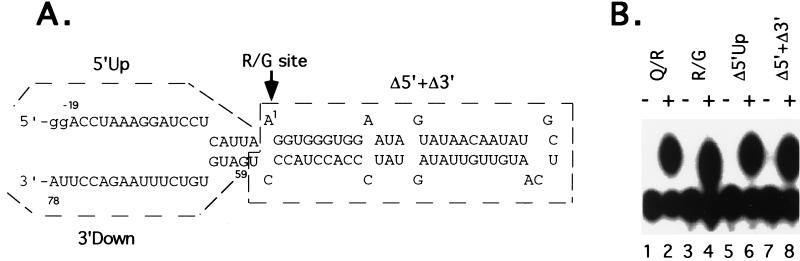
Figure 4.
Editing of the minimal R/G substrate for rRED1. (A) RNA sequence and predicted structure surrounding the GluR-B R/G site. The minimal R/G site RNA consists of 19 nucleotides upstream of the R/G site (5′Up), 58 nucleotides of R/G site plus ECS sequence (Δ5′ + Δ3′) and 20 nucleotides downstream of the ECS sequence (3′Down). A dinucleotide (gg) was added to its 5′ end for efficient transcription. Arrow indicates the adenosine of the R/G editing site. (B) Editing activity was determined in the absence (−) or presence (+) of rRED1 for different lengths of R/G substrates. Q/R site RNA is used as a control (Q/R, lanes 1, 2). The minimal R/G site RNA (R/G, lanes 3 and 4) was made by RNA ligation of unlabeled 21 mer RNA (pppGGACCUAAAGGAUCCUCAUUA) and 5′ end-labeled 78 mer RNA lacking the 5′Up sequence (Δ5′Up). Deletion of 5′Up (Δ5′Up, lanes 5 and 6) and deletion of 5′Up and 3′Down (Δ5′ + Δ3′, lanes 7 and 8) were made by in vitro transcription based on PCR-generated templates. The adenosine at the editing site was added by ApG dinucleotide during transcription and labeled using T4 polynucleotide kinase.