Abstract
The effects of culture conditions and chloramphenicol treatment on the induction of the marine bacterium Pseudoalteromonas spongiae to larval settlement of Hydroides elegans were investigated. The results showed that P. spongiae cells grown in the medium containing both yeast extract and peptone (YP-grown P. spongiae) was highly inductive to larval settlement, whereas P. spongiae cells grown in the medium containing only peptone (P-grown P. spongiae) or YP-grown P. spongiae cells treated with chloramphenicol at the onset of biofilm development (YPC-grown P. spongiae) did not induce larval settlement. Analysis of biofilm formation, biofilm structure, and the surface protein profile indicated that only the induction-capable YP-grown P. spongiae formed a well-developed biofilm, while the P-grown P. spongiae and the YPC-grown P. spongiae did not. We report here for the first time that bacterial biofilm formation was associated with its induction of larval settlement.
In the marine environment, natural and artificial substrata are readily colonized by micro- and macroorganisms in a process known as “biofouling” (4, 8, 52). The dioecious, free-spawning, tube-building polychaete Hydroides elegans (Haswell 1883) is one of the most troublesome fouling organisms, occurring widely in tropical and subtropical seawaters (41, 51). Larval settlement of H. elegans marks the turning point from a planktonic life stage to a sessile life stage and represents a crucial step in biofouling. Factors affecting larval settlement are therefore the focus of biofouling studies and antifouling control.
Competent larvae of H. elegans settled rapidly after induction by marine natural biofilms or certain monospecies bacterial films in the laboratory (26, 51). Larvae of H. elegans settle only in the presence of a metabolically active biofilm, however, not on a clean surface (21, 27, 51). Previous studies suggested that the bacterium-derived settlement cues were produced after bacteria attached to a surface and the cues were biofilm surface associated (11, 13, 19, 25). However, little attention was paid as to whether the bacterial biofilm formation was involved in bacterial induction of larval settlement. Bacteria undergo profound changes in physiological features during biofilm formation, a dynamic process wherein bacteria transform from planktonic (free-swimming) organisms to cells that are part of a complex, surface-attached community (5, 9, 15, 35, 36, 42, 47). Recent research revealed that many kinds of bacterial activity, such as infection or symbioses and production of bioactive compounds, were associated with biofilm formation (2, 12, 32, 53). Microbiologists have turned to biofilm formation for explanations of interesting microbial behaviors/bioactivities (37, 38).
In this study, employing the newly described marine bacterium Pseudoalteromonas spongiae (24, 28), we investigated the possible relevance of biofilm formation for its ability to induce larval settlement of H. elegans.
Effect of culture condition on bacterial induction of larval settlement.
In a preliminary experiment, bacterial culture conditions including temperature (16°C and 30°C), salinity (17 ppt and 34 ppt), yeast extract (0 g lliter−1 and 3 g liter−1), peptone (0 g liter−1 and 5 g liter−1), and glucose (0 g liter−1 and 10 g liter−1) were combined according to the Latin square design L8(27) in order to determine which factor(s) had a significant effect on bacterial biofilm's induction of larval settlement of H. elegans. Bacterial cells from different culture conditions were harvested from the broth by centrifugation (3,000 × g, 10 min), washed with autoclaved filtered seawater (AFSW), and then resuspended in AFSW (optical density at 600 nm of 0.15). Four milliliters of bacterial suspension was added to a polystyrene petri dish (diameter, 50 mm; Falcon no. 1006), incubated at 24°C for 3 h under a static condition to develop a single-layer bacterial film (26). Larval settlement bioassays were then performed according to references 26 and 39. The results showed that among the five factors, temperature, salinity, glucose, and peptone had no significant effect (P > 0.05), while yeast extract was the only factor having significant effect on the induction of the biofilm: P < 0.05; one-way analysis of variance (ANOVA); F = 77.65 > F0.05(1, 2) = 18.51.
To confirm the importance of yeast extract for the bacterial induction of larval settlement, Pseudoalteromonas spongiae was grown in media containing 8 g liter−1 yeast extract (Y) and peptone (P) at different ratios, namely, Y/P ratios of 0:8, 1:7, 3:5, 5:3, 7:1, and 8:0. Bacteria grew well in all media; bacterial biomasses in different media after cultivation were not significantly different from each other (one-way ANOVA; P < 0.05). However, biofilms of bacteria from different culture media showed remarkable difference in induction of larval settlement. P. spongiae cells grown in the medium containing only peptone (designated P-grown P. spongiae) or only yeast extract (designated Y-grown P. spongiae) showed no induction (19% of larvae settled) or only moderate induction (46% of larvae settled), respectively, whereas P. spongiae cells grown in media containing both Y and P (YP-grown P. spongiae), no matter at what ratio, were highly inductive to larval settlement (more than 63% of larvae settled). The highest settlement rate, 78%, was recorded in response to the bacterium grown in the medium with a 3:5 Y/P ratio (YP-grown P. spongiae). In all cases, bacterial densities in biofilms before the larval settlement bioassay were around 3 × 105 cells mm−2 and were not significantly different from each other (one-way ANOVA; P < 0.05).
These results indicated that presence of yeast extract in the culture medium was essential for the bacterial induction of larval settlement. Nutrient availability could be the cause for the difference, since peptone consists of only protein hydrolysis products, while yeast extract contains different types of nutrients such as trace metals, vitamins, etc. The key element in the yeast extract for the biofilm's inductivity deserves further investigation in the future.
Marine Pseudoalteromonas spp. are capable of producing bioactive compounds (18). Several studies of the relationship between marine Pseudoalteromonas spp. and invertebrate larval settlement suggested that Pseudoalteromonas spp. might provide settlement cues for invertebrate larvae (18, 20, 33). In our study, supernatants from planktonic culture and biofilm-conditioned seawater of the YP-grown P. spongiae and P-grown P. spongiae cells had no inductive effects, suggesting that the inductive cues should be surface-associated small compounds or macromolecules, which was consistent with previous studies (19, 25, 27).
Effect of chloramphenicol treatment on bacterial induction of larval settlement.
The highly inductive biofilm of YP-grown P. spongiae was treated with chloramphenicol (100 μg ml−1; Sigma) and tested for larval settlement. Chloramphenicol served here as the protein synthesis inhibitor (14). In a preliminary experiment, we found that incubating the bacterial cells in chloramphenicol (100 μg ml−1 in AFSW) for 3 h did not significantly (P < 0.05) reduce the number of viable bacteria (3-h-treated cultures, 1.06 × 108 CFU ml−1; untreated culture, 1.28 × 108 CFU ml−1), while in a disk diffusion assay, chloramphenicol did exert an inhibitive effect on protein synthesis, since no bacteria grew around the paper disk.
Development of bacterial films was performed the same as described before. Chloramphenicol was added at the onset (0 h) of biofilm formation or after 10 h of biofilm formation to a final concentration of 100 μg ml−1. In both cases, after 3 h of treatment, chloramphenicol solution was removed before larval settlement bioassay; parallel dishes without the chloramphenicol treatment served as controls. The larval settlement bioassay showed that chloramphenicol treatment at the onset of biofilm formation significantly reduced the biofilm's level of induction (one-way ANOVA; P < 0.05), and only 16% of larvae settled on the treated biofilm, whereas 72% of larvae settled on the control biofilm. In contrast, treatment at the 10th hour of biofilm formation did not affect the biofilm's ability to induce.
These results indicated that the initial 10 h of biofilm development was essential for the biofilm's ability to induce. O'Toole and Kolter reported that protein synthesis was required for initiation of biofilm formation in Pseudomonas fluorescens WCS365 (36). Treatment of chloramphenicol at the onset of biofilm formation might affect the protein synthesis or the activity of some regulatory proteins, which in turn would affect its biofilm development and produce induction cues for larval settlement, while after 10 h of development, those proteins that were required for presenting larval settlement induction might have already been synthesized (50), which in turn would maintain the biofilm's ability to induce larval settlement. The surface protein profiles of YP-grown P. spongiae and P-grown P. spongiae before attachment and after 10 h of attachment were then further investigated.
Bacterial surface protein profile.
Surface proteins were extracted by increasing the pH value (34) and analyzed in 12% separating gel using standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis protocols (23). After electrophoresis, protein bands were stained with Coomassie blue.
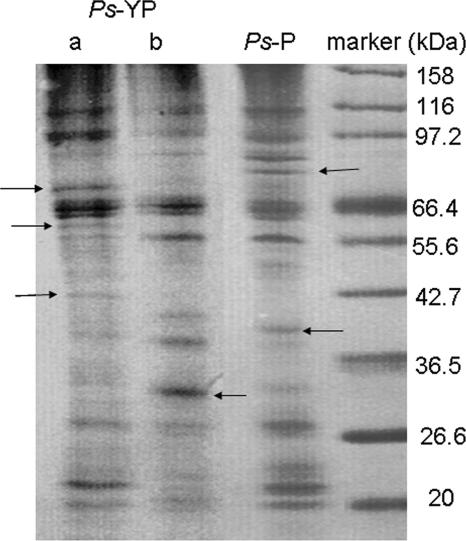
The results showed that the surface protein profile of YP-grown P. spongiae changed remarkably after attachment (Fig. 1). Some bands, such as those around 40 and 32 kDa in lane b, were unique in the YP-grown P. spongiae before attachment, whereas some bands, such as those around 70, 60, and 42 kDa in lane a, were unique in the YP-grown P. spongiae after attachment, suggesting that some specific surface proteins were either newly presented or shed during bacterial biofilm formation. It was also observed that some protein bands, such as bands around 97 and 22 kDa in lane a, were up-regulated, while some protein bands, such as bands around 55 and 38 kDa in lane b, were down-regulated after biofilm formation. These results indicated that the surface protein profile of YP-grown P. spongiae changed remarkably during the first 10 h of biofilm formation. Sauer et al. reported that more than 30 genes and 40 proteins were altered within 6 h following the attachment of Pseudomonas putida (44).
FIG. 1.
Surface protein profiles of YP-grown P. spongiae (Ps-YP) and P-grown P. spongiae (Ps-P) as analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Lane a, the surface protein profile after attachment; lane b, surface protein profile before attachment. Samples were analyzed in a 12% separation gel and stained with Coomassie blue. Arrows indicate the unique bands in each sample.
On the other hand, the surface protein profile of the P-grown P. spongiae did not change distinguishably after attachment. In comparison to the surface protein profile of the YP-grown P. spongiae before attachment, the P-grown P. spongiae did share some common protein bands, such as the bands around 116, 97, 66, and 56 kDa, representing the identical surface proteins for this bacterial species (Fig. 1). However, overall they were different, especially these bands around 70, 60, and 42 kDa: these bands, which appeared only in the YP-grown P. spongiae after attachment, did not appear in the P-grown P. spongiae, indicating that these proteins may be biofilm-specific proteins.
The “biofilm phenotype” is loosely defined by the patterns of protein and gene expression associated with biofilm cultures in comparison to those associated with planktonic culture (37). Therefore, an alteration in the protein profile is expected in bacteria after biofilm formation (43, 45). Here, we investigated only the surface protein profile, since surface proteins were considered to be important in the interaction between bacteria and surfaces (6, 16, 40) and only surface proteins could have any direct interaction with settling larvae. In the present study, the surface protein profile of the YP-grown P. spongiae changed remarkably after attachment, whereas changes in that of the P-grown P. spongiae were not distinguishable, suggesting that the YP-grown P. spongiae underwent a biofilm formation process, whereas the P-grown P. spongiae did not. Biofilm formation of YP-grown P. spongiae, YP-grown P. spongiae treated with chloramphenicol at the onset of biofilm formation (YPC-grown P. spongiae), and P-grown P. spongiae was further investigated.
Bacterial biofilm formation analysis.
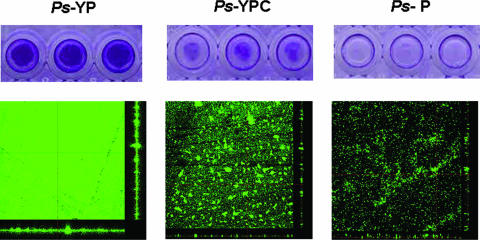
Analysis of bacterial biofilm formation was performed in a 96-well polystyrene plate (10). The results showed that the YP-grown P. spongiae cells attached well to the surface of the well, whereas a treatment with chloramphenicol significantly reduced the biomass of YPC-grown P. spongiae cells (Fig. 2). The P-grown P. spongiae cells could hardly attach to the surface of the well (Fig. 2). These results suggested that the P-grown P. spongiae was not capable of biofilm formation, whereas the YP-grown P. spongiae was capable of biofilm formation, yet chloramphenicol treatment inhibited its biofilm formation.
FIG. 2.
Biofilm formation of Pseudoalteromonas spongiae under different conditions. (Top panels) Biofilm formation analysis in 96-well polystyrene plates with three replicates. The extent of biofilm formation was determined by a crystal violet assay. (Bottom panels) CLSM images of biofilms. Biofilms were stained with fluorescein isothiocyanate-conjugated concanavalin A and were viewed at a ×400 magnification. Ps-YP, Pseudoalteromonas spongiae grown in medium containing yeast extract and peptone; Ps-YPC, YP-grown P. spongiae treated with chloramphenicol at the onset of biofilm formation; Ps-P, Pseudoalteromonas spongiae grown in medium containing peptone.
Biofilm structures of YP-grown P. spongiae, YPC-grown P. spongiae, and P-grown P. spongiae were further investigated by confocal laser scanning microscopy (CLSM). A bacterial suspension (optical density at 600 nm of 0.5) in AFSW was incubated in a petri dish (diameter, 90 mm; Sterilin, United Kingdom) containing a glass slide for 10 h in order to develop a biofilm. Biofilms were then stained with 1 mg ml−1 solution of fluorescein isothiocyanate-conjugated concanavalin A (Sigma) and visualized under a CLSM at a magnification of ×40 (dry, NA, 0.75; Nikon C1, Japan). Two replicate biofilms were formed for each treatment. For each biofilm, three image stacks were taken. In total, there were six image stacks for each treatment. The three-dimensional CLSM image of a biofilm was quantified using the computer program COMSTAT (15). Biofilm biomass, mean thickness, maximum thickness, surface coverage, and roughness were chosen to characterize the biofilm structure.
The CLSM pictures showed that biofilm structures of YP-grown P. spongiae (with or without chloramphenicol) and P-grown P. spongiae differed from each other (Fig. 2). Characterization of biofilm structures by COMSTAT (15) showed that the total biomass, substratum coverage, and maximum/mean thickness of the YP-grown P. spongiae biofilm were significantly higher (P < 0.05; t test) than those of the treated YP-grown P. spongiae biofilm and P-grown P. spongiae biofilm, whereas the roughness coefficient of the YP-grown P. spongiae biofilm was significantly lower (P < 0.05; t test) (Table 1). These values indicated that the YP-grown P. spongiae could effectively build up a biofilm with three-dimensional architecture in 10 h and tent to form a thick and even biofilm, while the chloramphenicol treatment inhibited biofilm development. As for the P-grown P. spongiae, few bacteria attached to the surface; the substratum coverage and mean thickness were very low, suggesting that the P-grown P. spongiae cells were unable to effectively attach to the surface and build up a biofilm.
TABLE 1.
Total biomass, maximum thickness, mean thickness, roughness coefficient, and substratum coverage of biofilms of YP-, YPC-, and P-grown P. spongiaea
| P. spongiae biofilm | Total biomass (μm3/μm2) | Thickness (μm)
|
Roughness coefficient | Substratum coverage | |
|---|---|---|---|---|---|
| Maximum | Mean | ||||
| YP grown | 6.94 ± 0.87 | 19.00 ± 2.35 | 8.03 ± 1.06 | 0.25 ± 0.02 | 0.99 ± 0.02 |
| YPC grown | 1.83 ± 0.09 | 7.60 ± 0.96 | 1.49 ± 0.20 | 0.74 ± 0.08 | 0.65 ± 0.05 |
| P grown | 0.76 ± 0.04 | 11.40 ± 1.19 | 0.99 ± 0.06 | 1.63 ± 0.03 | 0.11 ± 0.02 |
Values are means ± standard deviations of data from six CLSM image stacks.
A five-stage process is generally accepted to describe the development of biofilm formation. This process includes initial reversible surface attachment, irreversible attachment by producing extracellular polymeric substance (EPS), early development of biofilm architecture, maturation of biofilm architecture, and dispersion of single cells from the biofilm (47). The first three stages can happen in hours (35, 43). In the present study, biofilm formation of P. spongiae was performed under static conditions and for a short period of time (10 h) as compared to biofilm formation performed in a flow system. It is not surprising that no complex biofilm architecture such as the mushroom shape or water channels were observed in our biofilms (47). Our data on biofilm structure suggested that biofilm formation of the YP-grown P. spongiae had progressed to the third stage where bacteria attached irreversibly to the surface and started developing the biofilm architecture, whereas treatment with chloramphenicol arrested biofilm development at the first or second stage. On the other hand, biofilm formation in the P-grown P. spongiae was unlikely to happen. The bacterium seemed to stay at the first stage, where the reversibly attached cells were easily washed off in the 96-well plate analysis.
Bacterial biofilm formation is believed to be influenced by environmental signals and regulatory pathway (46). Nutrient availability is a common environmental signal that affects biofilm formation in different bacterial species. For example, phosphate, iron, and glucose were reported to affect biofilm formation of a Citrobacter sp., Pseudomonas aeruginosa, and Staphylococcus aureus, respectively (1, 3, 29). Molecular genetic analyses have also begun to reveal regulatory proteins and, through them, the environmental signals that affect biofilm formation (3, 29, 46). In this study, yeast extract in the medium provided necessary nutrients that enable YP-grown P. spongiae to develop a biofilm subsequently, whereas chloramphenicol treatment at the onset of biofilm formation might inhibit certain regulatory proteins and hence inhibit the biofilm formation.
The importance of bacterial biofilms in inducing larval settlement of certain benthic invertebrates in the marine environment has long been realized (11, 17, 22, 52). Previous studies on the interaction between a bacterial biofilm and a settling larva had pointed out that EPS of the biofilm might play an important role in inducing larval settlement (19, 25, 30, 31, 49). In fact, EPS also plays an important role in bacterial biofilm formation (7, 47, 48). EPS consists not only of polysaccharides but also considerable amounts of proteins, nucleic acids, and lipids (48). More detailed investigation of EPS with consideration of the protein activity would help to elucidate the mechanism underlying the induction of larval settlement by bacterial biofilm.
In conclusion, our data showed that the ability of the marine bacterium P. spongiae to induce larval settlement of H. elegans was associated with its biofilm formation. This study is the first one to investigate the effect of biofilm formation on bacterial induction of larval settlement. However, the question remains open as to how biofilm formation affects bacterial induction of larval settlement. A biofilm formation-deficient mutant would be necessary in the future to address this question.
Acknowledgments
We thank Arne Heydorn from Technical University of Denmark for kind assistance in running the COMSTAT program and Ching-Man Chan and Vivian Yu from the Biology Department of HKUST for help with using the confocal laser scanning microscope. We also thank Maris McEdward, On On Lee, Vengatesen Thiyagarajan, Hans-U. Dahms, Jaug-Seu Ki, and Jan Pechenik for valuable comments on the manuscript.
This investigation was supported by Hong Kong RGC grants (HKUST6402/05 M, CA04/05.Sc01, and COMAR07/08.Sc01) to P.-Y. Qian.
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Allan, V., M. Callow, E. Macaskie, and M. Paterson-Beedle. 2002. Effect of nutrient limitation on biofilm formation and phosphatase activity of a Citrobacter sp. Microbiology 148:277-288. [DOI] [PubMed] [Google Scholar]
- 2.Allegrucci, M., F. Z. Hu, K. Shen, J. Hayes, G. D. Ehrlich, J. C. Post, and K. Sauer. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 188:2325-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banin, E., M. Vasil, and P. Greenberg. 2005. Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. USA 102:11076-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooksey, K., and B. Wigglesworth-Cooksey. 1995. Adhesion of bacteria and diatoms to surfaces in the sea—a review. Aquat. Microb. Ecol. 9:87-96. [Google Scholar]
- 5.Costerton, J., Z. Lewandowski, D. Caldwell, R. Korber, and H. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 6.Cucarella, C., C. Solano, J. Valle, B. Amorena, Í. Lasa, and J. R. Penadós. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flemming, H. 2002. Biofouling in water systems—cases, causes and countermeasures. Appl. Microbiol. Biotechnol. 59:629-640. [DOI] [PubMed] [Google Scholar]
- 9.Fletcher, M. 1996. Bacterial attachment in aquatic environments: a diversity of surfaces and adhesion strategies, p. 1-24. In M. Fletcher (ed.), Bacterial adhesion: molecular and ecological diversity. Wiley, New York, NY.
- 10.Genevaux, P., S. Muller, and P. Bauda. 1996. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 30:27-30. [DOI] [PubMed] [Google Scholar]
- 11.Hadfield, M., and V. Paul. 2001. Natural chemical cues for the settlement and metamorphosis of marine invertebrate larvae, p. 431-461. In J. G. McClintock and B. J. Baker (ed.), Marine chemical ecology. CRC Press, Boca Raton, FL.
- 12.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 13.Harder, T., S. C. K. Lau, H. U. Dahms, and P. Y. Qian. 2002. Isolation of bacterial metabolites as natural inducers for larval settlement in the marine polychaete Hydroides elegans (Haswell). J. Chem. Ecol. 28:2029-2043. [DOI] [PubMed] [Google Scholar]
- 14.Hash, J. H. 1972. Antibiotic mechanisms. Annu. Rev. Pharmacol. 12:35-56. [DOI] [PubMed] [Google Scholar]
- 15.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 16.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905-918. [DOI] [PubMed] [Google Scholar]
- 17.Holmstrom, C., and S. Kjelleberg. 2000. Bacterial interactions with marine fouling organisms, p. 101-115. In L. V. Evans (ed.), Biofilms: recent advances in their study and control. Harwood Academic, Amsterdam, The Netherlands.
- 18.Holmstrom, C., S. Egan, A. Franks, S. McCloy, and S. Kjelleberg. 2002. Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol. 41:47-58. [DOI] [PubMed] [Google Scholar]
- 19.Huang, S. Y., and M. G. Hadfield. 2003. Composition and density of bacterial biofilms determine larval settlement of the polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 260:161-172. [Google Scholar]
- 20.Huggett, M. J., J. E. Williamson, R. de Nys, S. Kjelleberg, and P. D. Steinberg. 2006. Larval settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia 149:604-619. [DOI] [PubMed] [Google Scholar]
- 21.Hung, O. S., V. Thiyagarajan, R. S. S. Wu, and P. Y. Qian. 2005. Effect of ultraviolet radiation on biofilms and subsequent larval settlement of Hydroides elegans. Mar. Ecol. Prog. Ser. 304:155-166. [Google Scholar]
- 22.Kirchman, D., S. Graham, D. Reish, and R. Mitchell. 1982. Bacteria induce settlement and metamorphosis of Janua (Dexiospira) brasiliensis Grube (Polychaeta: Spirorbidae). J. Exp. Mar. Biol. Ecol. 56:153-163. [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lau, S. C. K., M. M. Y. Tsoi, X. C. Li, S. Dobretsov, Y. Plakhotnikova, P. K. Wong, and P. Y. Qian. 2005. Pseudoalteromonas spongiae sp nov., a novel member of the gamma-Proteobacteria isolated from the sponge Mycale adhaerens in Hong Kong waters. Int. J. Syst. Evol. Microbiol. 55:1593-1596. [DOI] [PubMed] [Google Scholar]
- 25.Lau, S. C. K., T. Harder, and P. Y. Qian. 2003. Induction of larval settlement in the serpulid polychaete Hydroides elegans (Haswell): role of bacterial extracellular polymers. Biofouling 19:197-204. [DOI] [PubMed] [Google Scholar]
- 26.Lau, S. C. K., K. K. W. Mak, F. Chen, and P. Y. Qian. 2002. Bioactivity of bacterial strains isolated from marine biofilms in Hong Kong waters for the induction of larval settlement in the marine polychaete Hydroides elegans. Mar. Ecol. Prog. Ser. 226:301-310. [Google Scholar]
- 27.Lau, S. C. K., and P. Y. Qian. 2001. Larval settlement in the serpulid polychaete Hydroides elegans in response to bacterial films: an investigation of the nature of putative larval settlement cue. Mar. Biol. 138:321-328. [Google Scholar]
- 28.Lee, O. O., and P. Y. Qian. 2003. Chemical control of bacterial epibiosis and larval settlement of Hydroides elegans in the red sponge Mycale adherens. Biofouling 19:171-180. [DOI] [PubMed] [Google Scholar]
- 29.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maki, J. S., L. Ding, J. Stokes, J. H. Kavouras, and D. Rittschof. 2000. Substratum/bacterial interactions and larval attachment: films and exopolysaccharides of Halomonas marina (ATCC 25374) and their effect on barnacle cyprid larvae, Balanus amphitrite Darwin. Biofouling 16:159-170. [Google Scholar]
- 31.Maki, J. S., and R. Mitchell. 1985. Involvement of lectins in the settlement and metamorphosis of marine invertebrate larvae. Bull. Mar. Sci. 37:675-683. [Google Scholar]
- 32.Murray, T. S., M. Egan, and B. I. Kazmierczak. 2007. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr. Opin. Pediatr. 19:83-88. [DOI] [PubMed] [Google Scholar]
- 33.Negri, A. P., N. S. Webster, R. T. Hill, and A. J. Heyward. 2001. Metamorphosis of broadcast spawning corals in response to bacteria isolated from crustose algae. Mar. Ecol. Prog. Ser. 223:121-131. [Google Scholar]
- 34.Ohlendieck, K. 1996. Extraction of membrane proteins, p. 293-304. In S. Doonan (ed.) Protein purification protocols. Humana Press, Totowa, NJ.
- 35.O'Toole, G., H. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 36.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 37.Parsek, M. R., and C. Fuqua. 2004. Biofilms 2003: emerging themes and challenges in studies of surface-associated microbial life. J. Bacteriol. 186:4427-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasmore, M., and J. W. Costerton. 2003. Biofilms, bacterial signaling, and their ties to marine biology. J. Ind. Microbiol. Biotechnol. 30:407-413. [DOI] [PubMed] [Google Scholar]
- 39.Pechenik, J. A., and P. Y. Qian. 1998. Onset and maintenance of metamorphic competence in the marine polychaete Hydroides elegans Haswell in response to three chemical cues. J. Exp. Mar. Biol. Ecol. 226:51-74. [Google Scholar]
- 40.Pratt, L. A., and R. Kolter. 1999. Genetic analyses of bacterial biofilm formation. Curr. Opin. Microbiol. 2:598-603. [DOI] [PubMed] [Google Scholar]
- 41.Qian, P. Y. 1999. Larval settlement of polychaetes. Hydrobiologia 402:239-253. [Google Scholar]
- 42.Sauer, K. 2003. The genomics and proteomics of biofilm formation. Genome Biol. 4:219-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauer, K., and A. K. Camper. 2001. Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J. Bacteriol. 183:6579-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Southey-Pillig, C. J., D. G. Davies, and K. Sauer. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187:8114-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52:917-924. [DOI] [PubMed] [Google Scholar]
- 47.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 48.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 49.Szewzyk, U., C. Holmstrom, M. Wrangstadh, M. O. Samuelsson, J. S. Maki, and S. Kjelleberg. 1991. Relevance of the exopolysaccharide of marine Pseudomonas sp strain S9 for the attachment of Ciona-intestinalis larvae. Mar. Ecol. Prog. Ser. 75:259-265. [Google Scholar]
- 50.Szomolay, B., I. Klapper, J. Dockery, and P. S. Stewart. 2005. Adaptive responses to antimicrobial agents in biofilms. Environ. Microbiol. 7:1186-1191. [DOI] [PubMed] [Google Scholar]
- 51.Unabia, C., and M. G. Hadfield. 1999. The role of bacteria in larval settlement and metamorphosis of the polychaete Hydroides elegans. Mar. Biol. 133:55-64. [Google Scholar]
- 52.Wieczorek, S. K., and C. D. Todd. 1998. Inhibition and facilitation of settlement of epifaunal marine invertebrate larvae by microbial biofilm cues. Biofouling 12:81-118. [Google Scholar]
- 53.Yan, L. M., K. G. Boyd, D. R. Adams, and J. G. Burgess. 2003. Biofilm-specific cross-species induction of antimicrobial compounds in bacilli. Appl. Environ. Microbiol. 69:3719-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]