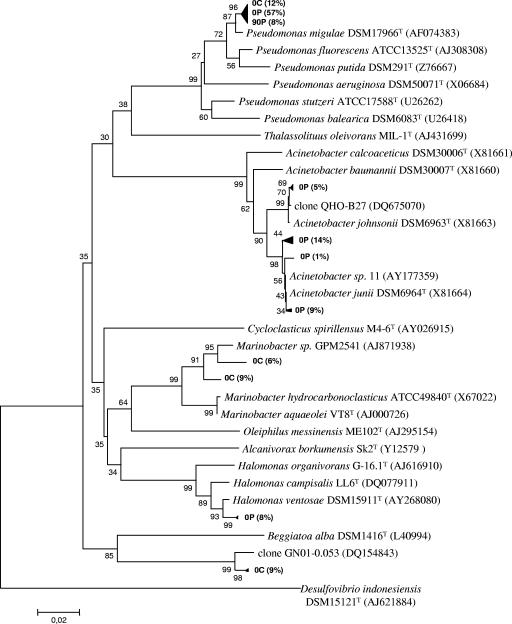
FIG. 5.
Phylogenetic tree based on the analysis of reverse-transcribed 16S rRNA cloned sequences (673 bp aligned) from control and contaminated microcosms sampled at the beginning of the experiment (0C and 0P, respectively) and a contaminated microcosm sampled after 90 days of incubation (90P). Only clones (in bold type) and their closest relative sequences affiliated with Gammaproteobacteria are shown. The numbers in parentheses indicate the percentage of clones (difference, ≤1 bp) in the corresponding microcosm library. The tree was rooted with 16S rRNA gene sequences of the strain Desulfovibrio indonesiensis DSM15121T. The scale bar corresponds to 0.02 substitutions per nucleotide position. Percentages of 100 bootstrap resamplings that supported the branching orders in each analysis are shown above or near the relevant nodes.