Abstract
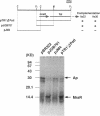
The mraR gene, which has a coding frame of 363 bp and lies close to and upstream of the ftsI gene of Escherichia coli, is involved in both cell division and cell lysis. It is thought to function in regulating the two distinct steps of the cell cycle, as two different one-base mutations in this unique gene caused different phenotypical changes in the cell. Comparison of nucleotide sequences of the mutant type mraR DNAs with the wild type suggested that filamentation of the cell was caused by a mutation in the putative start codon, whereas lysis of the cell was caused by a mutation which led to a change of one internal glutamate residue to lysine.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- DeVries J. K., Zubay G. DNA-directed peptide synthesis. II. The synthesis of the alpha-fragment of the enzyme beta-galactosidase. Proc Natl Acad Sci U S A. 1967 Apr;57(4):1010–1012. doi: 10.1073/pnas.57.4.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez M. J., Fluoret B., van Heijenoort J., Ayala J. A. Nucleotide sequence of the regulatory region of the gene pbpB of Escherichia coli. Nucleic Acids Res. 1990 May 11;18(9):2813–2813. doi: 10.1093/nar/18.9.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino F., Jung H. K., Ikeda M., Doi M., Wachi M., Matsuhashi M. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J Bacteriol. 1989 Oct;171(10):5523–5530. doi: 10.1128/jb.171.10.5523-5530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino F., Matsuhashi M. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem Biophys Res Commun. 1981 Aug 14;101(3):905–911. doi: 10.1016/0006-291x(81)91835-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Leclerc G., Noël G., Drapeau G. R. Molecular cloning, nucleotide sequence, and expression of shl, a new gene in the 2-minute region of the genetic map of Escherichia coli. J Bacteriol. 1990 Aug;172(8):4696–4700. doi: 10.1128/jb.172.8.4696-4700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M., Wachi M., Ishino F. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res Microbiol. 1990 Jan;141(1):89–103. doi: 10.1016/0923-2508(90)90101-u. [DOI] [PubMed] [Google Scholar]
- Nakamura M., Maruyama I. N., Soma M., Kato J., Suzuki H., Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191(1):1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Nishimura Y., Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci U S A. 1978 Feb;75(2):664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]