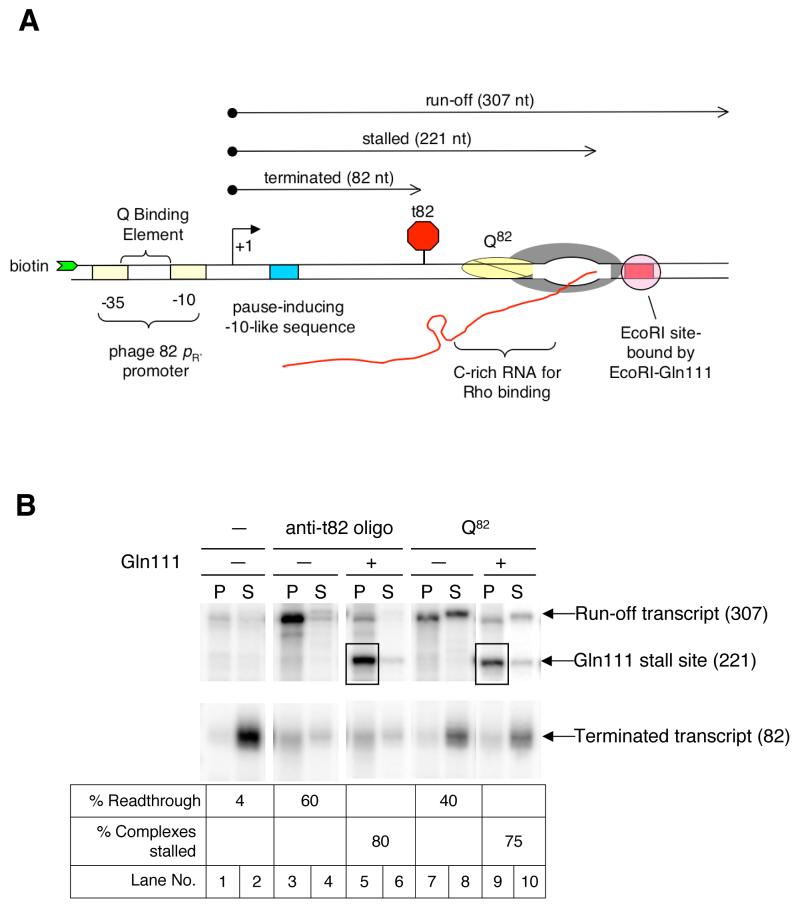
Figure 1.
- Schematic of the experimental method used to construct defined populations of Q82-modified complexes. The template was obtained from pSS100 and contained the phage 82 late gene promoter, the intrinsic terminator t82, a C-rich region for optimal Rho binding to the RNA, and an EcoR1 site. Distances are indicated from the transcription start site.
- Gel resolution of elongation complexes filtered past t82 using either the oligo anti-t82 or the antiterminator Q82, and stalled at a downstream site using the roadblock protein EcoR1-Gln111. After incubation with the indicated components, reaction mixtures were separated into magnetic bead-bound pellet (P) and supernatant (S) fractions. Numbers in parentheses indicate lengths of the RNA transcripts.