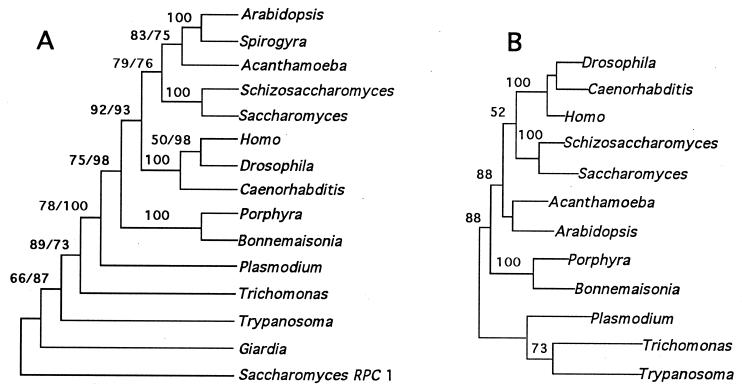
Figure 2.
Phylogenetic analyses based on RPB1 sequences. (A) Congruent bootstrapped parsimony (paup 3.1.1; ref. 24) and neighbor-joining (protdist, phylip3.53; ref. 25) trees. Trees are unrooted, but the largest subunit of polymerase III RPC1 from S. cerevisiae was used as the outgroup (21). Distances were estimated using the Dayhoff PAM matrix for amino acid substitutions. Percent appearances in 1,000 parsimony/100 distance bootstrap replicates are given above each node. Only one value is given when bootstrap values agree. (B) Tree with the maximum likelihood (codeml, paml 1.1; ref. 26) using the Jones, Taylor, Thornton matrix for amino acid substitutions. Branch lengths are approximately proportional to the estimated number of substitutions leading to the subsequent node. Six categories were used to estimate the γ distribution for rate variation, and likelihood estimates made with α = 1 and α = 0.5 produced the same tree topology. Because paml disregards any position in an alignment in which a gap occurs in any one of the aligned sequences, truncated sequences (Giardia, RPC1, Spirogyra) were deleted from the analysis to prevent loss of large portions of the data set.