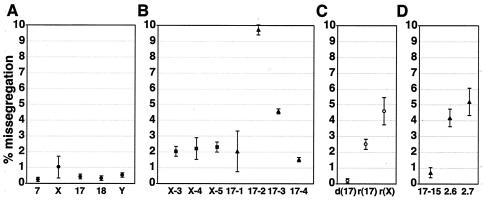
FIG. 3.
Missegregation of natural, artificial, and variant chromosomes. Missegregation rates were calculated as the number of missegregating chromosomes divided by the total number of chromosomes present of each type. Error bars represent one standard deviation from the mean of two segregation experiments for each chromosome. The total number of cells scored for each data point was >400. (A) Missegregation rates for natural chromosomes 7, X, 17, 18, and Y. (B) Missegregation rates for artificial chromosomes in cell lines X-3, X-4, X-5, 17-1, 17-2, 17-3, and 17-4. DXZ1-based artificial chromosomes (squares) and D17Z1-based artificial chromosomes (triangles) are indicated. (C) Missegregation frequencies for chromosome 17 with a deletion [d(17)], the ring chromosome 17 [r(17)], and the ring X chromosome [r(X)]. (D) Missegregation rates for D17Z1-based artificial chromosomes in cell lines 17-15, PF2.6, and PF2.7.