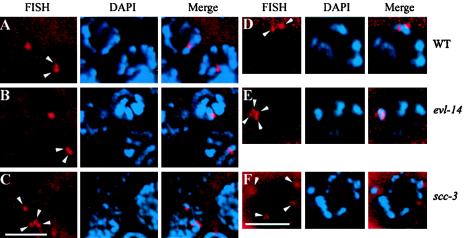
FIG. 5.
Detection of homologous pairing and sister chromatid cohesion by FISH and DAPI staining. (A to C) Nuclei in the pachytene zone of the germ line. Arrowheads indicate the FISH signals. In the wild type (A) and the evl-14 mutant (B), two closely located FISH signals in each nucleus were detected, representing paired homologs and normal sister chromatid cohesion. (C) In contrast, four separated signals were detected in the scc-3 mutant, indicating the lack of sister chromatid cohesion. (D to F) Oocytes at the diakinesis stage. (D) In the wild type, two FISH signals were observed. (E) In the evl-14 mutant, four closely located FISH signals are visible, suggesting that the sister chromatid cohesion is not as stable as that in the wild type. (F) In the scc-3 mutant, four completely separated FISH signals are visible (two of them slightly out of focus), which is consistent with the data presented for panel C. Bar, 5 μm.