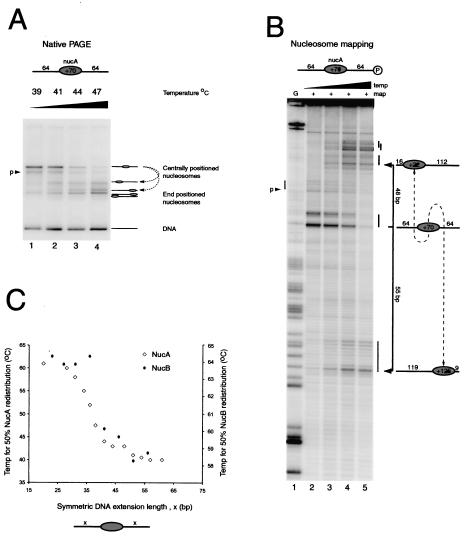
FIG. 1.
Increased linker DNA length reduces the temperature required for thermally driven nucleosome redistribution. (A) Native gel electrophoresis (PAGE) of nucleosomes assembled onto the 64A64 DNA fragment following incubation at 39, 41, 44, and 47°C (lanes 1 to 4, respectively). Most of the nucleosomes start as a discrete slowly migrating species, with a small proportion as a second species labeled “p.” Following thermal incubation, faster migrating species are generated. (B) Site-directed mapping of the same samples as for panel A (lanes 2 to 5, respectively). Nucleosomes generate a characteristic pattern of two cleavage sites separated by 7 bp and allow determination of nucleosome positions at base pair resolution using the G ladder in lane 1, as diagrammed to the right (nucleosomes are not to scale). A small amount of mapping is also observed at a second site labeled “p.” After thermal incubation, mapping products accumulate within 16 bp of either end of the DNA fragment. (C) Plot of the temperature required to shift 50% of nucleosomes from their initial central location on DNA fragments to the symmetric extensions of flanking DNA. Temperatures required for shifting on the MMTV NucA (open diamonds; left axis) and MMTV NucB (filled diamonds; right axis) positioning sequences are shown.