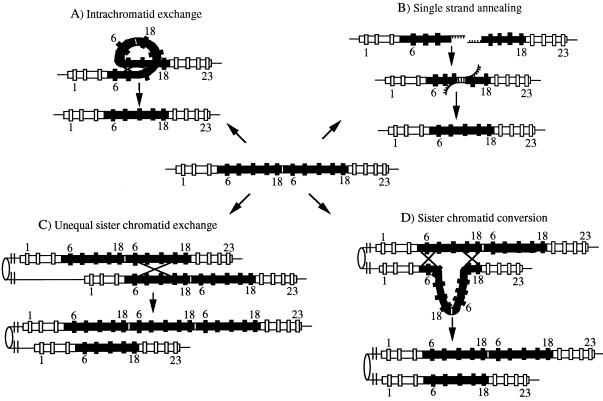
Figure 1.
pun structure and possible mechanisms of intrachromosomal recombination resulting in deletions (according to refs. 11, 17, and 20). In the center the pun structure is shown with exons 6–18 duplicated (21). (A) Intrachromatid crossing-over occurs after pairing of the two copies of the pun duplication in a looped configuration (11). Crossing over results in deletion of one of the two copies giving rise to reversion to p+. (B) Single-strand annealing is initiated by a double-strand break between the duplicated exons (20, 22); DNA ends are degraded by a 5′–3′ single-strand specific exonuclease to expose the flanking homologous sequences. Annealing of the complementary single strands occurs, and the nonhomologous ends are removed followed by DNA synthesis and ligation. (C) Unequal sister chromatid exchange occurs as crossing over between one copy of the exon duplication on one sister chromatid and the other copy of the exon duplication on the other sister chromatid. Reciprocal products are the deletion of one copy of the exon 6–18 duplication resulting in reversion to p+ and the triplication of exons 6–18. (D) Sister chromatid conversion events can occur after unequal pairing of the homologous portions of both copies of the exon duplication on one sister chromatid with either one of the two copies on the sister chromatid having the duplicated sequence in a looped-out configuration (11). Double crossover or gene conversion may lead to a conversion event during which one of the two copies of exons 6–18 is lost. The other sister chromatid maintains its original configuration. This event may also be initiated by a double-strand break between the duplicated copies on one sister chromatid degradation with a single-strand exonuclease until to the region of homology after which invasion, D-loop formation, and repair synthesis might happen from the sister chromatid (11).