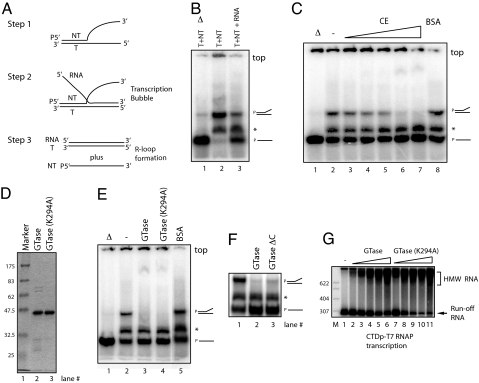
Fig. 3.
The GTase domain of human CE promotes RLF. (A) Experimental strategy for the RLF assay. Template strand (T), nontemplate strand (NT), and RNA are indicated. NT strand was labeled at its 5′ end (P). (B) RLF assay. Partially hybridized T and NT strands are shown (lane 2). Addition of RNA resulted in spontaneous RLF, converting ≈60% of NT into single-stranded NT (ssNT, lane 3). Heat-denatured products as molecular makers are shown in lane 1. Partially hybridized T and NT strands and ssNT are indicated on the right side. Asterisk indicates products made by aberrant T and NT hybridization. Top of the gel is indicated. (C) CE promotes RLF. RLF assays were performed with increasing amounts (0, 20, 50, 100, 200, and 400 ng) of CE (lanes 2–7) or 400 ng of BSA (lane 8). (D) Purified wild-type human GTase and GTase(K294A) (2 μg, respectively) were resolved by SDS/PAGE and visualized by Coomassie staining. (E) RLF assays were performed with 200 ng of wild-type human GTase (hGTase, lane 3), hGTaseK294A (lane 4), or BSA (lane 5). (F) RLF assays were performed with 200 ng of each GTase (lane 2) and GTase-active mutant having a C-terminal truncation of 28 aa (lane 3). (G) In vitro transcription with CTDp-T7 RNAP was performed with increasing amounts (30, 60, 90, 120, and 150 ng) of hGTase (lanes 2–6) or hGTaseK294A (lanes 7–11). Samples were resolved by nondenaturing PAGE in B, C, E, and F and by denaturing PAGE in G.