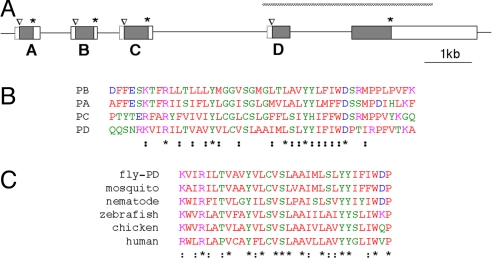
Fig. 4.
The inaF motif. (A) Diagram of the Drosophila genome from the inaF region showing additional exons that flank the exon for INAF-B. Shaded boxes delineate ORFs in the exons; the initial methionine and stop codon for each of them are marked with a caret and an asterisk, respectively. Open boxes enclosed by solid lines indicate UTRs deduced from the sequence of cDNAs. These cDNAs reveal that exons A–D are each spliced to the acceptor site of the rightmost exon; as a result, the methionine in this exon that was marked with a caret in Fig. 1A does not appear to function as the start of an ORF (it is an internal methionine of the inaF-D gene) and is now left unmarked. Open boxes enclosed by dashed lines indicate 5′ UTRs whose extent is uncertain. As in Fig. 1A, the extent of the P106x deletion is diagrammed with a hatched bar. (B) Alignment of a segment of the four ORFs diagramed in A. Residues are colored according to the Clustal (www.ebi.ac.uk/clustalw) convention. Asterisks and semicolons below the last sequence mark positions of identity or conservative substitution, respectively. (C) Alignment of the inaF motif found in INAF-D with similar motifs found in from ORFs from the indicated organisms. Residues are colored as in B, as are positions of conservation.