Abstract
Objective
The neural cell adhesion molecule (NCAM1) is a multifunction transmembrane protein involved in synaptic plasticity, neurodevelopment, and neurogenesis. Multiple NCAM1 proteins were differentially altered in bipolar disorder and schizophrenia. Single nucleotide polymorphisms (SNPs) in the NCAM1 gene were significantly associated with bipolar disorder in the Japanese population. Bipolar disorder and schizophrenia may share common vulnerability or susceptibility risk factors for shared features in each disorder.
Methods
Both SNPs and splice variants in the NCAM1 gene were analysed in bipolar disorder and schizophrenia. A case-control study design for association of SNPs and differential exon expression in the NCAM1 gene was used.
Results
A genotypic association between bipolar disorder and SNP b (rs2303377 near mini-exon b) and a suggestive association between schizophrenia and SNP 9 (rs646558) were found. Three of the two marker haplotypes for SNP 9 and SNP b showed varying frequencies between bipolar and controls (P < 0.0001) as well as between schizophrenia and controls (P < 0.0001). There were nine NCAM1 transcripts present in postmortem brain samples that involve alternative splicing of NCAM1 mini-exons (a, b, c) and the secreted (SEC) exon. Significant differences in the amounts of four alternatively spliced isoforms were found between NCAM1 SNP genotypes. In exploratory analysis, the c—SEC alternative spliced isoform was significantly decreased in bipolar disorder compared to controls for NCAM1 SNP b heterozygotes (P = 0.013).
Conclusions
Diverse NCAM1 transcripts were found with possibly different functions. The results suggest that SNPs within NCAM1 contribute differential risk for both bipolar disorder and schizophrenia possibly by alternative splicing of the gene. Psychiatr Genet 17:55−67 © 2007 Lippincott Williams & Wilkins.
Keywords: alternative splicing, association, bipolar disorder, genetics, neural cell adhesion molecule 1, schizophrenia, secreted-neural cell adhesion molecule 1, single nucleotide polymorphism, variable alternative spliced exon–secreted-neural cell adhesion molecule 1
Introduction
The neural cell adhesion molecule 1 (NCAM1) might be a candidate gene for both bipolar disorder (BPD) and schizophrenia (SZ) (Vawter, 2000). NCAM1 is found on chromosome 11q23.1 and is part of the immunoglobin superfamily. NCAM1 is expressed in both neurons and glial cells with cell recognition properties involved in cellular migration, synaptic plasticity (Ronn et al., 1998; Kiss and Muller, 2001) and central nervous system development (Ronn et al., 1998). Three major isoforms of NCAM1 [180, 140, 120 (Cunningham et al., 1987)] arise from a single copy gene via RNA processing, specifically alternative splicing and polyadenylation (Gower et al., 1988), whereas NCAM1, 105–115 kDa, appears to result from posttranslational modifications (Vawter, 2000). NCAM1 involves several alternatively spliced exons such as the variable alternative spliced exon (VASE) (Van Duijnhoven et al., 1992; Reyes et al., 1993) and the secreted exon (SEC) (Gower et al., 1988), which are expressed in brain and appear to be developmentally regulated (Vawter, 2000). Many other splice patterns have been identified in NCAM1 and if they were to be translated, it is predicted that they could result in up to 192 different NCAM1 proteins (Barthels et al., 1992).
In particular, the secreted isoform of NCAM1 (SECNCAM1) (Bock et al., 1987) is increased in the hippocampus of BPD patients (Vawter et al., 1999) whereas a proteolytic cleavage isoform of NCAM (cNCAM) was not altered in the brain of patients with BPD but increased in SZ (Vawter et al., 1999). Transgenic NCAM1 mice show some suggestive parallels to SZ such as alterations in brain ventricular ratio and alterations in prepulse inhibition (Wood et al., 1998). Additionally, a recent mouse model that overexpresses a truncated form of NCAM1 similar to the cleaved form of NCAM1 in human neuropsychiatric disorders also exhibits alterations in synaptophysin, GABAergic neurons, and behavior (Pillai-Nair et al., 2005).
Chromosome 11q has been an area of interest, as it harbors several candidate genes for neuropsychiatric disorders including the D2 dopamine receptor (DRD2) at 11q23 (Berry et al., 2003). NCAM1 and DRD2 are ∼146 kb apart from each other. Linkage studies (Maziade et al., 1995) and genome-wide scans (Levinson et al., 1998; Gurling et al., 2001; Lewis et al., 2003) have kept interest in the 11q area because the results suggest that this region contains loci that increase susceptibility to SZ. DRD2 genetic studies have often produced mixed results (Dubertret et al., 2004). A recent study, however, using both a case–control association study and a transmission disequilibrium test showed that two single nucleotide polymorphisms (SNPs) of DRD2 and a polymorphism of the novel X-Kinase gene which lies between NCAM1 and DRD2 are significantly associated with SZ (Dubertret et al., 2004). In addition, three SNPs of NCAM1 were significantly associated with BPD (Arai et al., 2004), renewing interest in the possible involvement of NCAM1 as a candidate gene.
The mechanisms that control alternative splicing of NCAM1 for SEC or VASE in human brain have not been studied and would be of direct interest to the NCAM1 hypothesis of neuropsychiatric disorders (Jorgensen, 1995; Poltorak et al., 1996; Ronn et al., 1997; Vawter, 2000). Studies of neuropsychiatric disorders and NCAM1 have predominantly focused on protein levels in human brain and cerebrospinal fluid (CSF) levels. NCAM1 protein was elevated in the CSF of patients with mood disorders (Poltorak et al., 1996). SEC-NCAM1 was measured in the hippocampus of controls, suicide victims, and patients with BPD or SZ by quantitative Western immunoblotting (Vawter et al., 1999). In BPD, but not in SZ, an increased SEC-NCAM1 115/108 kDa ratio was found compared with controls. Cytosolic NCAM1 (105–115 kDa) protein derived from proteolytic cleavage was measured in comparison to synaptophysin. The NCAM1 (105–115 kDa)/synaptophysin ratio was increased in the hippocampus of SZ, but not in the BPD (Vawter et al., 1999). Additionally, VASE–NCAM1 was increased in the prefrontal cortex and hippocampus of bipolar patients (Vawter et al., 1998). Thus, BPD and SZ patients showed specific altered expression of SEC-NCAM1 and VASE–NCAM1 in brain. These differences of NCAM1 protein expression among BPD and SZ in hippocampus, prefrontal cortex and CSF are consistent with involvement of NCAM1 in a distinct molecular neuropathology for each neuropsychiatric disorder as postulated by others (Cremer et al., 1994; Bouras et al., 2001).
The evidence of altered NCAM1 expression in both BPD and SZ is consistent with reports that some genes and regions are associated with both diseases. Family studies, twin studies, linkages studies, and gene studies all point to overlap between the disorders (Craddock et al., 2005). The genetic studies indicate that variations at the same loci often influence susceptibility to both SZ and BPD, for example, DISC1 and NGR1 (Craddock et al., 2005).
The complete genomic organization of human NCAM1 gene was determined by comparing complementary deoxyribonucleic acid (cDNA) and genomic sequences (Saito et al., 1994; Arai et al., 2004). In a follow up genetic study of NCAM1 and BPD, 11 SNPs were found by the resequencing of NCAM1 (Arai et al., 2004). This association study looked at distribution of genotypic, allelic, and haplotype variants in BPD and unipolar disorder. Three SNPs, IVS6_32T_C (SNP 5), IVS7_11G_C (SNP 6, rs686050), and IVS12_21C_A (SNP 9, rs646558), displayed significant associations with BPD in a Japanese sample (Arai et al., 2004). The SNP haplotype significantly associated with BPD was located in a large linkage disequilibrium (LD) block possibly spanning SNPs 1–10 analyzed by Arai et al. (2004). These results imply that genetic variations in or near NCAM1 could be linked to risks associated with BPD in a Japanese sample. We hypothesized that by genotyping two of the same SNPs, rs686050 (SNP 6 located near the VASE exon) and rs646558 (SNP 9) studied in the Japanese sample, we could test the association in a North American sample and determine whether the same SNPs were in LD. We genotyped bipolar, SZ, and control samples.
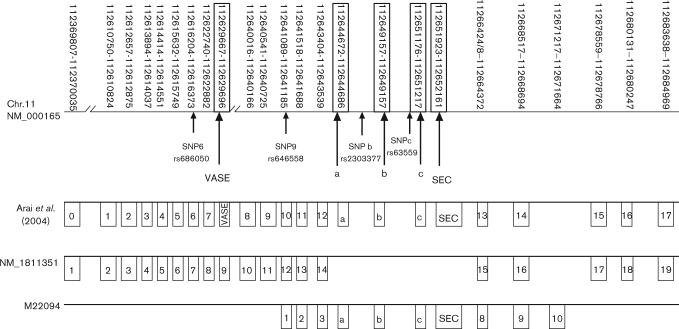
Between NCAM1 exons 12 and 13 (Fig. 1 Arai et al. Genomic Organization) is the secreted exon (SEC) and three mini-exons referred to as exon a, b, and c (Hamshere et al., 1991; Barthels et al., 1992; Arai et al., 2004). The most complex splicing patterns of NCAM1 are found in this region (Barthels et al., 1992). The role of these mini-exons (a, b, and c) in brain tissue is unknown. The splicing of the mini-exons in NCAM1 was studied in mouse muscle and neural tissue. In mouse muscle, exon ‘a’ was present in low levels, and exons ‘b’ and ‘c’ were usually incorporated together in conjunction with exon ‘a’. Mouse neural cells showed very low levels of exon ‘a’ and did not show any inclusion of exons ‘b’ and ‘c’ (Hamshere et al., 1991). Although the reading frame is maintained in all combinations of the mini-exon splice variants, it is not known whether the isoforms encode functional proteins. These mini-exons could be alternatively spliced to form tissue-specific NCAM1 isoforms and hence change the conformation of the extracellular region of the molecule. Exon ‘a’ may cause a ‘hinge’ in the molecule that could alter posttranslational modification (Hamshere et al., 1991). It is thought that alternative splicing of the mini-exons along with the larger exons may be key events that play signaling roles in embryogenesis and histogenesis (Prediger et al., 1988).
Fig. 1.
Neural cell adhesion molecule 1 (NCAM1) genomic organization and location of four polymorphic sites. The gene spans 214 kb, but does not contain any known exonic single nucleotide polymorphisms (SNPs) in dbSNP (NCBI, Human Genome Build 34). The arrows indicate the location of the four polymorphisms and the five exons used in this exploratory analysis. Accession numbers are listed for three sequences. The Arai et al. (2004) sequence is numbered according to their resequencing results.
The SEC exon is spliced out when mRNAs with transmembrane domain coding potential are generated. When the SEC exon is spliced in, it results in a truncated form of NCAM1 with various properties. SEC-NCAM1 could function as a soluble modulator of intercellular adhesion and other cell–cell interactions, or it might function as part of the extracellular matrix and be involved in cell–substratum interactions (Gower et al., 1988; Moran and Bock, 1988). Therefore, one focus of this study was related to the alternative splicing of SEC and mini-exons in the NCAM1 RNA molecule. Here, we show a survey of SEC mRNA to determine which different splice variants exist in human brain.
Novel splice variants of NCAM1 mini-exons a, b, c, and exon SEC were studied in human brain from bipolar, unipolar, and control brain samples in relationship to genotypes of four SNPs in NCAM1. We hypothesized that a relationship between the alternative splicing of the exons in NCAM1 mRNA might be related to SNPs located near mini-exon ‘b’ (SNP b: rs635596) and mini-exon ‘c’ (SNP c: rs2303377).
Methods
DNA extraction and single nucleotide polymorphism analysis (sequencing and TaqMan)
Genomic DNA (gDNA) was extracted from human postmortem brain cerebellum tissue (Table 1) following a Trizol (Invitrogen, Carlsbad, California, USA) extraction and ethanol precipitation protocol. Primers were designed for SNP 9 (Table 2) using Primer Express software (ABI, Applied Biosystems, Foster City, California, USA) and then tested via PCR to determine correct band size. Using the SNP 9 primers, gDNA of 40 cases [20 controls, 9 BPDs, and 11 major depressive disorders (MDDs)] was sequenced with both the forward and reverse primers. The presence and absence of SNP 9 was detected from the sequencing results. A Taqman Genotyping Assay (ABI) was also used for SNP 9 (Assay ID: c_2998872_20) and allele calls were made using the Allelic Determination program (ABI Prism 7000 sequence detection system, Applied Biosystems). The results of genotyping from sequencing and TaqMan (Applied Biosystems) were compared for verification. There was complete concordance between sequencing and TaqMan methods for SNP 9 with the exception of one sample, which showed an ambiguous sequencing trace. Therefore, genotyping of the remaining three SNPs (SNP 6: c_613361_20, SNP b: c_2998870_1, and SNP c: c_2998868_10) was carried out on the ABI Prism 7000 sequence detection system using the predesigned kits from ABI listed above. The SNPs were located using SNP Browser 2.0 (Applied Biosystems) and searching NCAM1 genomic alignments to find exons a, b, and c and then choosing a SNP that was within a few base pairs of the 5′ end of the exon. SNPs b and c are intronic and found just before exons ‘b’ (7 bp upstream) and ‘c’ (12 bp upstream), respectively. Exon ‘a’ did not have a SNP in close proximity (Fig. 1).
Table 1.
Demographics of cases used for genotyping and expression
| Gender | Age | |||
|---|---|---|---|---|
| Source | Tissue | Diagnosis | (M/F) | Avg (StDev) |
| Genotyping | ||||
| Conte-Pritzker (DNA) | Postmortem-CB | Control (n=20) | 13/7 | 52.6 (16.5) |
| BPD (n=9) | 6/3 | 63.9 (17.7) | ||
| MDD (n=11) | 7/4 | 50.7 (14.8) | ||
| Total (n=40) | 26/14 | 52.6 (16.5) | ||
| NIMH (DNA) | Lymphocyte | BPD (n=26) | 10/16 | 45.5 (9.3) |
| Postmortem-DLPFC | ||||
| Stanley Foundation | Control (n=35) | 26/9 | 42.5 (7.6) | |
| Samples (DNA) | BPD (n=35) | 17/18 | 45.3 (10.5) | |
| SZ (n=35) | 26/9 | 42.6 (8.5) | ||
| Total (n=105) | 69/36 | 44.0 (8.9) | ||
| Expression | ||||
| Conte-Pritzker (cDNA) | Postmortem-DLPFC | Control (n=13) | 11/2 | 48.0 (17.6) |
| BPD (n=8) | 5/3 | 52.0 (17.9) | ||
| MDD (n=10) | 7/3 | 48.5 (13.51) | ||
| Total (n=31) | 23/8 | 49.2 (17.7) | ||
| Genotype Analysis | ||||
| Total Samples | Control (n=55) | 39/16 | 47.6 (12.1) | |
| BPD (n=70) | 33/37 | 51.6 (12.5) | ||
| SZ (n=35) | 26/9 | 42.6 (8.5) | ||
| Total (n=160) | 98/62 | 47.3 (11.0) |
BPD, bipolar disorder; DLPFC, dorsalateral prefrontal cortex; F, female; M, male; MDD, major depressive disorder; NIMH, National Institute of Mental Health; SZ, schizophrenia.
Table 2.
Primers for each DNA segment to probe possible combination of splice variants (exons a, b, c, SEC, and VASE), as well as for an exon outside of the splice sites for measuring total NCAM1
| Primer | Forward | Reverse |
|---|---|---|
| VASE | 5′-GACCCCATTCCCTCCAT CAC-3′ | 5′-GGCTACGCACCAC CATGTG-3′ |
| Exon a | 5′-GACGCAGCCAGTCCA TAGC-3′ | a |
| Exon b | 5′-CGTCTACCCCTGTTC CATTGTC-3′ | 5′-TCTGGTGGAGACAATG GAACAG-3′ |
| Exon c | 5′-TCCTGCCCTTGCAACCA 3′ | 5′-GGTTGCAAGGGCAG GAAGA-3′ |
| SEC exon | 5′-CCAAGCTGGTCTTCA TAATGCTCTA-3′ | 5′-TTTGATGCTTGAACACTAT GAACATG-3′ |
| Exon 3 | 5′-GGCGGCGCTCAATGG-3′ | b |
| Exon 8 | c | 5′-GATCAGGTTCACTTTAATA GAGTTTCCA-3′ |
| SNP9 for sequencing | 5′-CGCAGCCAGTCCGTAAG TAAAG-3′ | 5′-AAGCTGGACCGGCTAC TAGGA-3′ |
The numbering is shown in Fig. 1 according to accession M22094. SNP 9 was also genotyped by direct sequencing.
NCAM1, neural cell adhesion molecule 1; VASE, variable alternative spliced exon.
Only the forward primer could be designed because exon a is 14 bp.
Exon 3 is before the variable exons and only the forward primer was needed to PCR outside the exons. The reverse primer was designed within another exon.
Exon 8 is after the variable exons and only the reverse primer was needed to PCR outside the exons. The forward primer was designed within another exon.
The genotypes were collected on an additional 26 bipolar genomic DNA samples extracted from lymphocytes from the National Institute of Mental Health (NIMH) for all four SNPs: SNP 6, SNP 9, SNP b, and SNP c (see Fig. 1). A third cohort was genotyped consisting of the Stanley Foundation (Chevy Chase, Maryland, USA) 105 dorsalateral prefrontal cortex (DLPFC) microarray samples (n = 35 controls, n = 35 BPD, n = 35 SZ) (Table 1). The Stanley samples were genotyped for SNP 9 and SNP b. In the final analysis, the three groups of bipolar cases were combined and three control groups were merged and used for statistical comparisons of SNP 9 and SNP b.
Brain collection, RNA extraction, and cDNA synthesis
Human postmortem brain tissue (Table 1) was previously acquired with consent from the decedents' next-of-kin. The details of the collection procedures and scoring of agonal factors for each case was reported earlier (Tomita et al., 2004). Total RNA was extracted from human postmortem DLPFC brain tissue using Trizol (Invitrogen) and converted into cDNA using Taqman Reverse Transcription Reagents (Applied Biosystems). DLPFC cDNA from control (n =13), bipolar (n =8), and MDD (n =10) cases were analyzed for splice variants involving mini-exons a, b, c and the SEC exon. The 31 DLPFC samples were rated before-hand for agonal factor score (AFS) (Tomita et al., 2004) and included AFS = 0 (n =24), AFS=1 (n =5), and AFS=3 (n = 2). In the final results and statistical analysis only the AFS = 0 cases were included. A patient with an AFS = 0 is free of agonal factors such as coma, hypoxia, pyrexia, seizures, dehydration, hypoglycemia, multiple organ failure, head injury, and ingestion of neurotoxic substances and is also not associated with prolonged death (agonal duration less than 1 h) (Tomita et al., 2004).
Primer design, PCR, band purification, and sequencing
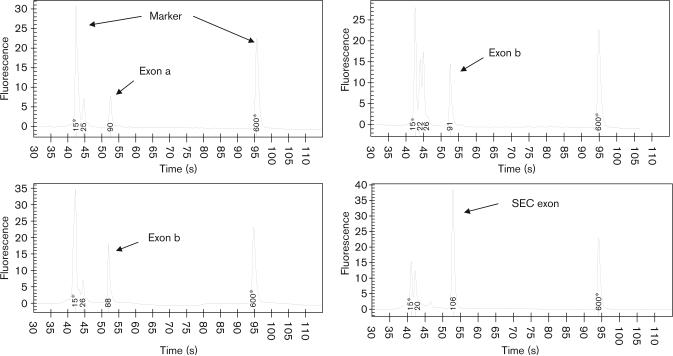
Primers were designed for each cDNA segment (Table 2) and possible combination of splice variant (a, b, c, and SEC) exons using Applied Biosystems Primer Express software (ABI). All PCR was performed on cDNA derived from human postmortem DLPFC. Initially, each single exon was amplified and then analyzed on an Aglient 2100 Bioanalyzer using the DNA 500 LabChip Kit (Agilent Technologies, Santa Clara, California, USA) to confirm the correct base pair size (Fig. 2). The PCR reaction was also visualized by agarose gel electrophoresis and the band of correct size was purified using the GFX PCR DNA and Gel Band Purification Kit (Amersham Biosciences, Pittsburgh, Pennsylvania, USA) and subsequently sequenced to prove that the correct exon was amplified.
Fig. 2.
Electropherograms of exons a, b, c, and secreted (SEC) cDNA following PCR and separated on Agilent 2100 Bioanalyzer. The identity of each peak was verified by direct sequencing.
Primers were then utilized in various combinations to amplify each possible splice variant that might be present and the Agilent 2100 Bioanalyzer (Agilent Technologies) was used to quantitate peak height and areas.
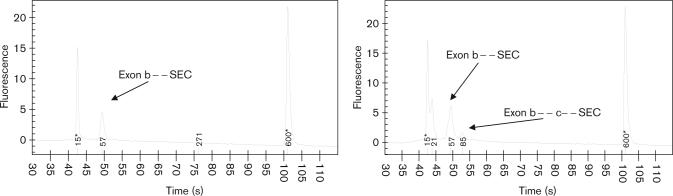
All of the individual exons were confirmed for sequence and base pair size. Thus, when the PCR was run using various combinations of the mini-exon primers, we were able to deduce which splice variant was present by examining the appropriate base pair reading on the electropherogram from the DNA LabChip (Agilent Technologies). For example, if we used the forward primer for exon ‘b’ and the reverse primer for the SEC exon we would examine the base pair peaks to determine whether the peaks included exon ‘b’ and the SEC exon and whether a larger base pair peak was present we could deduce that it included exon ‘c’ as well (Fig. 3). This process was carried out for all possible combinations of mini-exons and SEC.
Fig. 3.
Determination of splice variants of cDNA separated on the Agilent Bioanalyzer. PCR used the forward primer for exon b and the reverse primer for the secreted (SEC) exon. Each graph shows a different patient. The graph on the left shows one peak that corresponds to the splice variant b-SEC on the basis of base pair estimates of exon b and exon SEC. The graph on the right shows two peaks. One corresponds to the b-SEC splice variant and the other is slightly larger in base pairs. This peak is the b-c-SEC splice variant.
Quantitative reverse transcription-polymerase chain reaction using SybrGreen
The total amounts of SEC, NCAM1, and VASE were determined by quantitative real time PCR (Q-PCR) reactions. Primers were designed within the VASE and SEC exons and outside of the variable exons, but within an NCAM1 exon (Table 2). A template sample was selected for the standard curve with a concentration that ranged from 20 ng/μl to 6.3 pg/μl in three-fold dilution series. All reactions were 25 μl in total volume and carried out with SybrGreen PCR master mix from Applied Biosystems using primers designed as stated above. Reactions were completed in triplicate on the ABI Prism 7000 sequence detection system (Applied Biosystems) with the absolute quantitation program. Quantity of SEC, VASE, and NCAM1 was determined from the results and then normalized to the housekeeping gene protein phosphatase 1-alpha.
Statistical analysis
Hardy–Weinberg Equilibrium (HWE) was determined using XL Genetics (XLent Works, Warrnambool VIC, Australia) for the bipolar, control, and SZ cases. Departures from Hardy–Weinberg (DHW, http://hg-wen. uchicago.edu/dhw2.html) were analyzed using the DHW software (Wittke-Thompson et al., 2005) run via Mathematica 5.1 (Wolfram Research, Inc., Champaign, Illinois, USA). The DHW software tests for a goodness-of-fit for a locus showing departure from HWE. We ran the general model and derived the χ2 to test for goodness-of-fit. If the sample is a good fit then the underlying genetic disease model for a susceptibility locus in that region is a likely explanation for the DHW (Wittke-Thompson et al., 2005). Genotypic association was evaluated using the DeFinetti program (DeFinetti-Diagram and Hardy–Weinberg Test, Bonn, Germany, http://ihg.gsf.de/cgi-bin/ hw/hwa1.pl) for SNP 9 and SNP b. We checked for LD between SNP 9 and SNP b using Linkage Disequilibrium Analyzer 1.0 (Chinese National Human Genome Center, Beijing, China). The haplotype frequencies were estimated using the Estimate Haplotype (EH) Frequencies (Terwilliger and Ott, 1994) and EHplus (Zhao et al.., 2000) programs via the case–control analysis. The individual log-likelihood of the bipolar and the individual log-likelihood of the controls were each subtracted from the combined bipolar and control log-likelihood and the χ2 was derived.
Results
Allelic association
In a North American sample we did not replicate SNP 9 association with BPD that was previously reported in the Japanese sample (Arai et al., 2004). The allelic frequencies for bipolar and control cases were not different for SNPs b and 9 (Table 3). The odds ratio for SNP 9 allelic frequency was in accord with the prior study results of 1.5 for postmortem samples from the 40-cerebellum gDNA and the NIMH lymphocyte samples. Combining genotype data, however, with the bipolar samples from three sources (cerebellum gDNA, DLFC gDNA, and lymphocyte gDNA) the allelic effect was no longer present as the odds ratio dropped to 0.82.
Table 3.
Genotypic and allelic distributions for controls, bipolar disorder and schizophrenia cases
| Polymorphism | n | Genotype counts (frequency) | Allele counts (frequency) | P (Fisher's exact test) | |||
|---|---|---|---|---|---|---|---|
| SNP 9 | C/C | C/A | A/A | C | A | ||
| Control | 55 | 33 (0.60) | 22 (0.40) | 0 (0) | 88 (0.80) | 22 (0.20) | |
| BPD | 70 | 47 (0.67) | 23 (0.33) | 0 (0) | 117 (0.84) | 23 (0.16) | 0.466 |
| SZ | 35 | 24 (0.69) | 8 (0.23) | 3 (0.09) | 56 (0.80) | 14 (0.20) | 1 |
| BPD + SZ | 105 | 71 (0.68) | 31 (0.30) | 3 (0.02) | 173 (0.82) | 37 (0.18) | 0.602 |
| SNP b | T/T | T/C | C/C | T | C | ||
| Control | 55 | 16 (0.29) | 29 (0.53) | 10 (0.18) | 61 (0.55) | 49 (0.45) | |
| BPD | 70 | 19 (0.27) | 47 (0.67) | 4 (0.06) | 85 (0.61) | 55 (0.39) | 0.402 |
| SZ | 35 | 13 (0.37) | 19 (0.54) | 3 (0.09) | 45 (0.64) | 25 (0.36) | 0.24 |
| BPD + SZ | 105 | 32 (0.30) | 66 (0.63) | 7 (0.07) | 130 (0.62) | 80 (0.38) | 0.264 |
Fisher's exact P-values are shown for allelic distribution between case–controls.
BPD, bipolar disorder; SNP, single nucleotide polymorphism; SZ, schizophrenia.
With SNP b the combined results produced an odds ratio of 0.82 for BPD although the Fisher's exact was not significant (Table 3). The odds ratio of SZ was not significant for either SNP b or SNP 9.
Hardy–Weinberg equilibrium
The distributions of genotypes in loci SNP 9 were consistent with HWE for all groups and combinations of bipolar, controls, and schizophrenic groups as seen in the Japanese study (Arai et al., 2004). The exploratory SNP b was in HWE for all groups except the bipolar alone, controls plus bipolar, and control plus SZ plus bipolar. The departure from Hardy–Weinberg (DHW) in this sample for SNP b was tested for the genetic model of disease susceptibility as opposed to genotyping error or chance. Using the DHW software we tested the general model for the goodness-of-fit to the best-fit model and the result showed that the sample did not significantly differ from the goodness-of-fit (χ2 = 0.284, d.f. = 1, P = 0.5967). Therefore, the observed DHW is most likely due to the underlying genetic disease model.
Genotypic association
SNP 9 showed a suggestion of association with SZ, but not with BPD. The confidence interval (CI) for the odds ratio (OR) of SZ spanned above and below 1 (Table 4). The SNP 9 heterozygote [P = 0.010, OR 0.05, 95% CI (0.003–1.16)] and homozygote [P = 0.027, OR 0.08, 95% CI (0.004–1.67)] associations were significant, as well as allele positivity [P = 0.049, OR 9.57, 95% CI (0.47–193.92)] for SZ.
Table 4.
Genotypic association results
| Tests for genotypic association (risk allele 1) | ||||||
|---|---|---|---|---|---|---|
| Heterozygous (A/A ↔ C/A) | Homozygous (C/C ↔ A/A) | Allele positivity [(C/C + C/A) ↔ A/A] | ||||
| OR (CI) | χ2(P-value) | OR (CI) | χ2(P-value) | OR (CI) | χ2(P-value) | |
| SNP 9 | ||||||
| SZ | 0.054 (0.003−1.16) | 6.6 (0.01) | 9.57 [0.47−193.92] | 3.86 (0.049) | 0.084 [0.004−1.67] | 4.88 (0.027) |
| SNP b | C/C ↔ T/C | T/T ↔ C/C | (T/T + T/C) ↔ C/C | |||
| BPD | 4.05 (1.16−14.12) | 5.33 (0.02) | 2.97 (0.78−11.30) | 2.65 (0.103) | 3.66 (1.08−11.30) | 4.81 (0.028) |
SNP b showed heterozygote association with BPD [P = 0.021, OR 4.05, 95% CI (1.16–14.12)] and allele positivity [P = 0.028, OR 3.66, 95% CI (1.08–12.42)]. SNP b was not associated with SZ. The confidence interval for the OR of SZ spanned above and below 1 (Table 4).
Linkage disequilibrium
SNP 9 and SNP b were not in LD for control, BPD, or SZ and because of the small n of the current data set LD was explored further. We examined the SNP data at www.ensembl.org and used the CEPH (Utah residents with ancestry from Northern and Western Europe) population from HapMap (The International HapMap Project, www.hapmap.org) and also the Coriell Cell Repository population (European American descent) from Perlegen. The earlier study of Japanese samples observed LD between SNP 6 and SNP 9. Both populations, however, from the Ensembl of European ancestry showed that SNP 6 and SNP 9 were not in LD and also, SNP 9 and SNP b were not in LD. Additionally, SNP b and SNP c were not in LD although they are in close physical proximity.
Haplotype frequencies
Four haplotypes were formed with SNP 9 and SNP b alleles. For the EHplus program, haplotypes were formed based on genotype rather than alleles; there were eight possible haplotypes from the two SNPs (Sasieni, 1997). The distribution of all eight haplotypes was not significantly different between bipolar and controls or between SZ and controls. Continuing to explore the haplotypes we removed five haplotypes (n = 25) with low genotype frequencies (0.06–0.09) from the bipolar versus control group. The low frequencies were based on the genotype; however, the frequencies by allele were lower (∼0.03) for the haplotypes removed. The remaining three haplotypes (n = 100) were recalculated with the EHplus program (Zhao et al., 2002) (χ2 = 21.99, d.f. = 2, P = < 0.0001) and were significant after Bonferroni correction. The same analysis was performed on the SZ versus control group and again the top three haplotypes were significant (χ2 = 28.04, d.f. = 3, P = < 0.0001) and remained significant after Bonferroni correction. Bipolar plus SZ versus control was run and the same top three haplotypes were significant (χ2 = 27.90, d.f. = 2, P = < 0.0001) and also passed Bonferroni. For a final analysis we observed that SZ versus bipolar also showed significant differences in the same three haplotypes (χ2 = 16.41, d.f. = 2, P = 0.0003) (Table 5). This evidence suggests that the three most frequent haplotypes (by genotype) of SNP 9 and SNP b were distributed in different proportions between bipolar and controls as well as between SZ and controls. The ORs and haplotype frequencies suggest that there is a difference between bipolar, SZ, and controls at these two loci (Table 5).
Table 5.
SNP 9 and SNP b haplotype frequency (on the basis of genotype), odds ratio, and P-values
| Haplotype | Frequency (OR) | ||
|---|---|---|---|
| SNP 9-b | Control | BPD* | SZ# |
| C-T | 0.2 | 0.19 (0.95) | 0.31 (1.54) |
| C-(T/C) | 0.31 | 0.46 (1.48) | 0.37 (1.19) |
| (C/A)-(T/C) | 0.22 | 0.21 (0.95) | 0.11 (0.50) |
P-values were calculated from the χ2 values derived from the EHplus program.
BPD, bipolar disorder; OR, odds ratio; SNP, single nucleotide polymorphism; SZ, schizophrenia.
BPD vs. C P (value) <0.0001
SZ vs. C P (value) <0.0001; SZ vs. BPD P (value) 0.0003.
Neural cell adhesion molecule 1 splice variants
There are 16 possible splice variants involving four exon combinations (a, b, c, and SEC) in the NCAM1 molecule. Although not every splice variant was detected (Table 6), we found at least nine variants of these in human brain RNA. The individual patient's splice variant data are presented in Table 6. The patterns of individual variants are instructive from two perspectives, first they appear in certain combinations but not others, and second the combinations inherently lead to either the inclusion or the exclusion of the SEC exon. If the SEC exon is included the result is a premature termination of translation. This leads to a secreted NCAM1 molecule capable of acting as a ligand to many other molecules including NCAM1 and other cell recognition molecules. From the apparent combinations that involve SEC or splice SEC out of the RNA (Table 6) we were able to deduce these splicing rules for the mini-exons:
Table 6.
NCAM 1 splice variants from cDNA DLPFC
| a–b | a–c | a–b—c | a–SEC | b–c1 | b–SEC2 | a–c—SEC | b–c—SEC | c–SEC3 | |
|---|---|---|---|---|---|---|---|---|---|
| C1 | X | X | X | X | X | X | X | X | |
| C2 | X | X | X | X | X | X | X | ||
| C3 | X | X | X | X | X | X | X | ||
| C4 | X | X | X | X | X | ||||
| C5 | X | X | X | X | |||||
| C6 | X | X | X | X | X | X | X | ||
| C7 | X | X | X | X | X | X | X | ||
| C8 | X | X | X | X | X | X | |||
| C9 | X | X | X | X | X | X | |||
| C10 | X | X | X | X | X | X | X | ||
| C11 | X | X | X | X | X | X | X | ||
| C12 | X | X | X | X | X | X | X | X | |
| C13 | X | X | X | X | X | X | X | ||
| BPD1 | X | X | X | X | X | X | |||
| BPD2 | X | X | X | X | X | X | X | ||
| BPD3 | X | X | X | X | X | X | X | ||
| BPD4 | X | X | X | X | X | X | X | ||
| BPD5 | X | X | X | X | |||||
| BPD6 | X | X | X | X | X | X | X | ||
| BPD7 | X | X | X | X | X | X | X | ||
| BPD8 | X | X | X | X | X | X | X | ||
| MDD1 | X | X | X | X | X | X | X | ||
| MDD2 | X | X | X | X | X | X | X | ||
| MDD3 | X | X | X | X | X | X | X | ||
| MDD4 | X | X | X | X | X | X | X | ||
| MDD5 | X | X | X | X | X | X | X | X | |
| MDD6 | X | X | X | X | X | X | |||
| MDD7 | X | X | X | X | X | X | |||
| MDD8 | X | X | X | X | X | X | X | ||
| MDD9 | X | X | X | X | X | X | |||
| MD- | X | X | X | X | X | X | |||
| D10 |
Three splicing rules are described (in results). For example, rule 1 was derived from the three-numbered variants: 1b–c, 2b–SEC, and 3c–SEC which shows that b or c is always associated with SEC.
BPD, bipolar disorder; C, control; MDD, major depressive disorder; X, after PCR amplification, the splice variant was detected on the Agilent 2100 Bioanalyzer using the DNA 500 LabChip.
(1) NCAM1 SEC exon is spliced in 100% of the transcripts with mini-exon ‘b’.
(2) NCAM1 SEC exon is spliced in 100% of the transcripts with mini-exon ‘c’.
(3) NCAM1 SEC exon is spliced out 90–100% of transcripts with the introduction of mini-exon ‘a’ to either mini-exon ‘b’ or mini-exon ‘c’.
Thus, the splicing of mini-exon ‘b’ or ‘c’ produces a truncated NCAM1 protein that can be secreted.
Only cases with no agonal factors as defined in the methods were used for further quantification of splice variant results. We first examined all of the participants and found that the presence of agonal factors led to significant differences in gene expression when analyzing the splicing isoforms. Five variants were significantly different between AFS = 0 and AFS ≥ 1 participants: a–b (P = 0.01), a–c (P = 0.005), a–b–c (P = 0.003), b–c (P = 0.03), and c–SEC (P = 3.0E-06). Thus, participants with agonal factors were eliminated from further analysis.
Psychotropic drug use was explored in the bipolar and major depression cohorts to determine whether the drugs influenced the expression of NCAM1. Medical records revealed that all bipolar patients on lithium had discontinued use before death, ranging from days to months. Additionally, lithium was not detected in brain tissue assays. No significant differences were observed in the splice variant amounts between patients with lithium and patients without lithium. All patients with a history of prescribed selective serotonin reuptake inhibitor class of antidepressants at the time of death (BPD = 1 and MDD = 5) were compared with patients not prescribed at the time of death and there were no significant splice variant differences between the groups.
Genotype × splice variant differences
As splicing variations for NCAM1 followed certain combinations of mini-exons, but not others, it was possible that polymorphic variations near the splice junction for exons a, b, and c may be a factor in determining splicing patterns. The four NCAM1 SNPs were analyzed for an association with overall differences in NCAM1 expression levels, as well as the splice variants of the SEC and VASE exons.
Both VASE ( + ) and VASE ( – ) splice variant amounts were decreased in bipolar compared with control for both SNP b and SNP c genotype comparisons (Table 7). The difference in VASE–NCAM1 by SNP b and SNP c genotype is of interest because the presence of exon ‘b’ and exon ‘c’ potentially modifies NCAM1 secretion. Varying amounts of SEC splice variants exist in three of the four SNP genotypes (Table 7 and Fig. 4). Thus, from what we could identify, the amount of SEC-NCAM1 present was not regulated by one factor alone. The amount of the alternative spliced variant c–SEC shows significant differences between SNP b genotypes. Specifically, the amount of c–SEC mRNA was significantly increased in the heterozygous SNP b genotype compared with SNP b homozygote. Furthermore, the amount of SEC total measured by variant analysis was shown to be increased in the SNP 9 major allele homozygote (Table 7).
Table 7.
Genotype × splice variant differences
| SNP 6 G→C | SNP 9 C→A | SNP b T→C | SNP c G→A | |||
|---|---|---|---|---|---|---|
| Splice variant | G/C vs. C/C | C/C vs. C/A | T/T vs. T/C | T/C vs. C/C | T/T vs. C/C | G/G vs. G/A |
| Patients | (n=12 vs. n=11) | (n=16 vs. n=7) | (n=7 vs. n=10) | (n=10 vs. n=7) | (n=7 vs. n=7) | (n=5 vs. n=19) |
| a–SEC | 0.015 ↑ | |||||
| b–SEC | 0.016 ↓ | |||||
| SECa | 0.016 ↑ | |||||
| a–b | 0.043 ↑ | |||||
| c–SEC | 0.012 ↑ | |||||
| VASE (−) | 0.034 ↓ | 0.037 ↓ | ||||
| VASE (+) | 0.051 ↓ | 0.045 ↓ | ||||
The P-values are from t-test comparisons and arrows indicate direction of change relative to the first genotype in each comparison. For each of the four SNPs the genotypes were compared for splice variant amount by t-test to determine if the amount varied on the basis of genotype.
SNP, single nucleotide polymorphism; VASE, variable alternative spliced exon.
SEC is a measure of all of the splice variants that contained the SEC exon. The PCR was designed to span only the single exon.
Fig. 4.
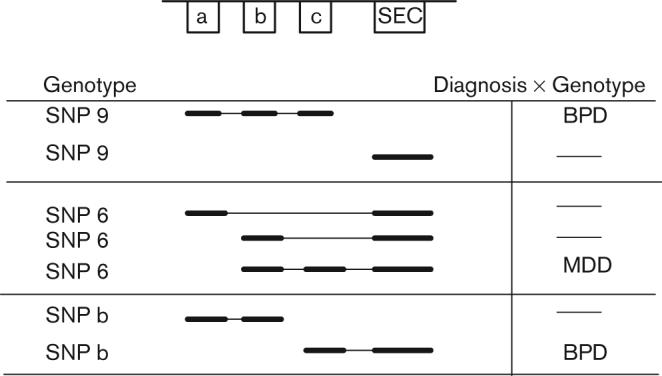
Significant alterations of neural cell adhesion molecule 1 (NCAM1) exon splice variant levels are shown for genotype and diagnosis by genotype.
The relative abundance of controls was assessed by genotype to see whether the splice variant differences seen in the total samples were also observed in the controls alone. There were no significant differences between genotypes for all four SNPs in controls.
The genotypes of the four SNPs and the splice variants with significant differences from the analyses of Genotype × Splice Variant (Table 7) were then categorized by diagnosis for exploratory analyses. Several splice variants showed a significant effect (Table 8).
Table 8.
Exploratory three-way analysis of genotype, splice variant, and diagnosis
| SNP | Genotype | Splice variant | BPD vs. C | MDD vs. C |
|---|---|---|---|---|
| SNP 9 | C/C | a–b–c | 0.052 ↑ | |
| SNP 6 | G/C | b–c–SEC | 0.050 ↑ | |
| SNP b | T/T | VASE (–) | 0.056 ↑ | |
| SNP b | T/C | c–SEC | 0.013 ↑ | |
| SNP b | T/C | NCAM1 Ct Q–PCR | 0.053 ↑ |
Significant genotype × splice variant differences × diagnosis (P-values) are shown.
For each SNP genotype and splice variant the splice variant amounts were evaluated by t-test on the basis of diagnosis and significant P-values and trends are shown.
Figure 4 displays a graphical summary of these findings.
NCAM1, neural cell adhesion molecule 1; SNP, single nucleotide polymorphism; VASE, variable alternative spliced exon.
SNPs 6, 9, and b each showed significant differences in splice variants that included the mini-exons and the SEC exon (Fig. 4). Each of these three SNPs had one significant variant that also varied by diagnosis (Table 8 and Fig. 4). For example, BPD patients heterozygous for SNP b showed a decrease of the c–SEC isoform compared with control heterozygotes (P < 0.013). The common link in many of these exploratory analyses was that each SNP was associated with differences in the amounts of SEC exon (Fig. 4).
Quantitative-reverse transcription-PCR
The relative amounts of SEC, VASE, and NCAM1 were determined by Q-PCR and examined by diagnosis. The raw Ct values were normalized with the housekeeping gene protein phosphatase 1-alpha and then evaluated by one-way analysis of variance. The bipolar group showed a significant decrease in the amount of VASE mRNA compared with controls. In addition, SNP b showed a diagnosis effect for NCAM1 measured by Q-PCR when comparing the heterozygotes. No differences were observed in the total amounts of NCAM1 or SEC by Q-PCR between genotypes (Table 8).
Discussion
The present data relate NCAM1 polymorphic variation to BPD and splice variations in mRNA occurring near the polymorphisms. A genotypic association exists between NCAM1 SNP b and BPD and a suggestive association of SNP 9 with SZ. Three of the two marker haplotypes for SNP 9 and SNP b, CT, C(T/C), and (C/A)(T/C) display varying frequency distribution between bipolar and controls as well as between SZ and controls. SNP b and SNP 9 are not in LD and they are individually related to SZ (SNP 9) and BPD (SNP b). The odds ratios show that BPD differs from SZ for SNP 9 and SNP b by haplotype frequency differences.
The allelic frequency for SNP 9 does not show an over transmission of the major allele in BPD that was shown in the Japanese study (Arai et al., 2004). This variation may be the result of population differences because SNP 9 and SNP 6 are not in LD in the North American sample. SNP 9 and SNP 6 were previously shown to be in LD among the Japanese population (Arai et al., 2004). The lack of LD in our samples was confirmed by calculation of LD for Caucasian populations listed on the Ensembl website for SNP 9 and SNP 6. Additional explanations for the different outcomes could be that our study missed the bipolar association with SNP 9 owing to a smaller sample size and power. This seems unlikely as the observed OR of SNP b (from 2.97 to 4.05) for BPD was larger than the SNP 9 OR (1.64) reported for BPD by Arai et al. (2004). Thus, intronic SNP b appears to be relevant to risk for BPD in samples of European ancestry.
The splice variant evidence for SNPs 9 and b confirm that each SNP can be associated with differences in SEC exon splicing, thus providing some differential mechanisms for the release of NCAM1 in the brain. We observed notable differences between polymorphisms in NCAM1 and the relative isoform variants of the SEC exon which can lead to truncation and secretion of NCAM1 in the brain (Gower et al., 1988). This finding concerning the difference in splice variant relative amounts as a function of certain genotypes is shown in three of the four SNPs in which at least one genotype shows a difference in the amount of SEC by splice variant. This evidence suggests that the amount of SEC-NCAM1 in brain is not regulated by just one genotype. As the haplotypes composed of SNP 9 and SNP b (by genotype) are significantly different between controls and bipolar and between controls and SZ this may support the observation of differential splicing patterns of the SEC exon found across many samples. Additionally, SNP 9 and SNP b are not in LD and thus the individual associations in SZ and bipolar with these SNPs may also be transmitted through differential splicing patterns.
The SEC exon was clearly regulated by certain combinations of mini-exons. Thus, although we do not know the mechanism that produces differences in the amount or splicing of SEC (Barthels et al., 1992), we have identified discrete splice variants that can be further studied and are perhaps associated with regulatory intronic SNPs. The major isoforms seen in all samples were exons b–c, b–SEC, and c–SEC. Certain mini-exon combinations were not detected, such as the a–b–c–SEC, indicating that certain mechanisms which influence the pattern of splicing have yet to be discovered.
In addition to the SEC splicing observations, we also found connections between the amount of VASE involving the SNP b and c genotypes. VASE is significantly decreased in bipolar compared with controls by Q-PCR. It is also decreased in the minor homozygote of SNP b and the heterozygote of SNP c by variant analysis. This further strengthens the use of polymorphism and splice variant comparisons to gain a better understanding of how NCAM1 is processed.
Our findings of the genotypic association of SNP b with BPD along with association of the same polymorphisms with NCAM1 gene expression in brain present an interesting and perhaps fundamental mechanism that might ultimately influence pathophysiology. One possible role of intronic SNP b in BPD is shown. First, patients heterozygous for SNP b or with allele positivity for the major allele show an increased risk for BPD. Thus, the major allele is present more often in bipolar than in controls. Second, the heterozygote for SNP b showed a greater amount of c–SEC compared with the minor allele homozygote for all patients. The bipolar SNP heterozygotes, however, showed a decrease in c–SEC compared with control heterozygotes. Hence, the increased risk for bipolar with the heterozygous SNP b and the decreased amount of c–SEC in the heterozygote leads to an overall decrease in SEC in BPD. A decrease in SEC spliced NCAM1, which produces a truncation of NCAM1, presumably leads to alterations in SEC-NCAM1 protein, consistent with previous findings in which SEC-NCAM1 protein levels were measured in BPD brain (Vawter et al., 1999). This three-way interaction between × diagnosis genotype × splice variant leads to alterations in the total amount of NCAM1 released.
The genotypic association for SNP b might be spurious as the bipolar sample was not in HWE. Thus, the deviation from HWE could be driving the association. The goodness-of-fit test indicates that this is probably not the case. The association is more likely driven by the genetic disease model for susceptibility and the departure from HWE is not due to chance or genotyping error.
The suggestive genotypic association of SNP 9 with SZ implies that over transmission of the major homozygous allele leads to an increased risk of SZ. These data also suggest that the heterozygote will be found more often in controls than in SZ. The confidence interval is very wide for this sample; therefore, we believe that a larger sample would be needed to confirm this association. We do not have splice variant data for the SZ samples, but we know that SNP 9 and SNP b are not in LD and the haplotype frequencies vary between SZ and controls. Further exploration of the splice variants in SZ would help to explain the suggestive genotypic association.
The role of NCAM1 and its susceptibility in both bipolar and SZ supports the idea of an overlap between the disorders in perhaps genetics and pathophysiology. By understanding how the diseases overlap there will be a better grasp of the classification of SZ and bipolar and furthermore the pathophysiology will be made clearer which will lead to improved treatments (Craddock et al., 2005).
Taken together, the simplified splicing rules and associations with genotypes promote a rich variation in the amount of secreted NCAM1 available in the brain. It is thought that secreted NCAM1 functions similarly to proteolytically cleaved NCAM1 (Vawter, 2000) in that nontethered NCAM1 can easily bind to other extracellular molecules such as tenascin (Rundus et al., 1998; Zacharias et al., 1999), L1 (Davis and Bennett, 1994), and receptors such as the FGF receptor (Williams et al., 1995; Saffell et al., 1997; Niethammer et al., 2002). All of these molecules can form assemblies between different cells and guide cellular migration, neurogenesis (Seki and Arai, 1995; Ní Dhúill et al., 1999; Casadesus et al., 2005), synaptic processes such as long term potentiation (Muller et al., 1996, 2000; Cremer et al., 1998; Bukalo et al., 2004; Dityatev et al., 2004), and learning (Cremer et al., 1994, 1997; Odumeru et al., 1997; Stork et al., 1997, 1999; Bukalo et al., 2004). These NCAM1 properties might be important mediators of neuropsychiatric illness (Barbeau et al., 1995; Poltorak et al., 1996; Vawter, 2000; Pillai-Nair et al., 2005).
We detected differences in two alternatively spliced exons of NCAM1 (VASE and SEC). In addition, we found that alternative splicing of mini-exons a, b, and c occurs in human brain and is presumably associated with a secreted NCAM1 protein, which can be confirmed in vitro. This exploratory analysis of association of NCAM1 splice variants in the human brain with polymorphisms can encourage further study to determine whether the causes for the differential expression can be made clearer. Our evidence of associations of two NCAM1 SNPs in bipolar and SZ and splice variant differences suggest that the rich variation in gene splicing might be one mechanism that promotes differences between SZ and BPD within the same gene.
Acknowledgements
We appreciate the assistance of Chief Deputy Coroner Jacque Berndt and staff of the Orange County Sheriff-Coroner's Office; as well as Kathleen Burke, Claudia Cervantes, and Karen Lopez. F. Warren Lovell, MD, who performed a neuropathological evaluation of the postmortem brains. Tissue specimens obtained from the Human Brain and Spinal Fluid Resource Center, VA West Los Angeles Healthcare Center, Los Angeles, CA 90073, which is sponsored by NINDS/NIMH, National Multiple Sclerosis Society, and Department of Veterans Affairs under the direction of Wallace W. Tourtellotte, MD, PhD.
We acknowledge members of the Pritzker Neuropsychiatric Disorders Research Consortium for providing DNA and RNA for this study: Kevin Overman, David Walsh, PsyD, Preston Cartagena, PsyD, Richard A. Stein, PhD, Hiro Tomita, MD, PhD, Steven G. Potkin, MD, and William E. Bunney Jr, MD (Department of Psychiatry and Human Behavior, College of Medicine, University of California, Irvine, California, USA) Jun Li, PhD and Rick Myers, PhD (Stanford Human Genome Center and Department of Genetics, Stanford University School of Medicine, Stanford, California, USA), Prabha Choudary, PhD and Edward G. Jones, MD (Center for Neurosciences University of California, Davis, Davis, California, USA), Stanley J. Watson, MD, PhD, Huda Akil, PhD, and Simon Evans, PhD (Mental Health Research Institute, University of Michigan, Ann Arbor, Michigan, USA). Members of the Pritzker Neuropsychiatric Disorders Research Consortium are supported by the Pritzker Neuropsychiatric Disorders Research Fund L.L.C. A shared intellectual property agreement exists between this philanthropic fund and the University of Michigan, the University of California, and Stanford University to encourage the development of appropriate findings for research and clinical applications.
We thank William F. Byerley (Department of Psychiatry, VA Medical Center, San Francisco, California, USA) for providing the NIMH bipolar lymphocyte DNA.
DNA was donated by The Stanley Medical Research Institute's brain collection courtesy of Drs Michael B. Knable, E. Fuller Torrey, Maree J. Webster, Serge Weis, and Robert H. Yolken.
The research was funded by the William Lion Penzner Foundation (UCI Department of Psychiatry) and by the NIMH Conte Center Grant P50 MH60398. NIMH bipolar lymphocyte DNA was collected through an NIMH grant (WFB).
References
- Arai M, Itokawa M, Yamada K, Toyota T, Arai M, Haga S, et al. Association of neural cell adhesion molecule 1 gene polymorphisms with bipolar affective disorder in Japanese individuals. Biol Psychiatry. 2004;55:804–810. doi: 10.1016/j.biopsych.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Barbeau D, Liang JJ, Robitalille Y, Quirion R, Srivastava LK. Decreased expression of the embryonic form of the neural cell adhesion molecule in schizophrenic brains. Proc Natl Acad Sci USA. 1995;92:2785–2789. doi: 10.1073/pnas.92.7.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthels D, Vopper G, Boned A, Cremer H, Wille W. High degree of NCAM diversity generated by alternative RNA splicing in brain and muscle. Eur J Neurosci. 1992;4:327–337. doi: 10.1111/j.1460-9568.1992.tb00880.x. [DOI] [PubMed] [Google Scholar]
- Berry N, Jobanputra V, Pal H. Molecular genetics of schizophrenia: a critical review. J Psychiatry Neurosci. 2003;28:415–429. [PMC free article] [PubMed] [Google Scholar]
- Bock E, Edvardsen K, Gibson A, Linnemann D, Lyles JM, Nybroe O. Characterization of soluble forms of NCAM. FEBS Lett. 1987;225:33–36. doi: 10.1016/0014-5793(87)81126-2. [DOI] [PubMed] [Google Scholar]
- Bouras C, Kövari E, Hof PR, Riederer BM, Giannakopoulos P. Anterior cingulate cortex pathology in schizophrenia and bipolar disorder. Acta Neuropathol. 2001;102:373–379. doi: 10.1007/s004010100392. [DOI] [PubMed] [Google Scholar]
- Bukalo O, Fentrop N, Lee A, Salmen B, Law JWS, Wotjak CT, et al. Conditional ablation of the neural cell adhesion molecule reduces precision of spatial learning, long-term potentiation, and depression in the CA1 subfield of mouse hippocampus. J Neurosci. 2004;24:1565–1577. doi: 10.1523/JNEUROSCI.3298-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesus G, Shukitt-Hale B, Stellwagen HM, Smith MA, Rabin BM, Joseph JA. Hippocampal neurogenesis and PSA–NCAM expression following exposure to 56Fe particles mimics that seen during aging in rats. Exp Gerontol. 2005;40:249–254. doi: 10.1016/j.exger.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Craddock N, O'Donovan MC, Owen MJ. The genetics of schizophrenia and bipolar disorder: dissecting psychosis. J Med Genet. 2005;42:193–204. doi: 10.1136/jmg.2005.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, et al. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- Cremer H, Chazal G, Goridis C, Represa A. NCAM is essential for axonal growth and fasciculation in the hippocampus. Mol Cell Neurosci. 1997;8:323–335. doi: 10.1006/mcne.1996.0588. [DOI] [PubMed] [Google Scholar]
- Cremer H, Chazal G, Carleton A, Goridis C, Vincent JD, Lledo PM. Long-term but not short-term plasticity at mossy fiber synapses is impaired in neural cell adhesion molecule-deficient mice. Proc Natl Acad Sci USA. 1998;95:13242–13247. doi: 10.1073/pnas.95.22.13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham BA, Hemperly JJ, Murray BA, Prediger EA, Brackenbury R, Edelman GM. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science. 1987;236:799–780. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- Davis JQ, Bennett V. Ankyrin binding-activity shared by the neurofascin/ L1/NRCAM family of nervous-system cell-adhesion molecules. J Biol Chem. 1994;269:27163–27166. [PubMed] [Google Scholar]
- Dityatev A, Dityateva G, Sytnyk V, Delling M, Toni N, Nikonenko I, et al. Polysialylated neural cell adhesion molecule promotes remodeling and formation of hippocampal synapses. J Neurosci. 2004;24:9372–9382. doi: 10.1523/JNEUROSCI.1702-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret C, Gouya L, Hanoun N, Deybach JC, Ades J, Hamon M, et al. The 3′ region of the DRD2 gene is involved in genetic susceptibility to schizophrenia. Schizophr Res. 2004;1:75–85. doi: 10.1016/s0920-9964(03)00220-2. [DOI] [PubMed] [Google Scholar]
- Gower HJ, Barton CH, Elsom VL, Thompson J, Moore SE, Dickson G, et al. Alternative splicing generates a secreted form of N-CAM in muscle and brain. Cell. 1988;55:955–964. doi: 10.1016/0092-8674(88)90241-3. [DOI] [PubMed] [Google Scholar]
- Gurling H, Kalsi G, Brynjolfsson J, Sigmundsson T, Sherrington R, Baljinder S, et al. Genome wide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q 32.2, 5q 33.2 and 8p 21–22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3–24 and 20q 12.1–11.23. Am J Hum Genet. 2001;66:661–673. doi: 10.1086/318788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamshere M, Dickson G, Eperon I. The muscle specific domain of mouse N-CAM: structure and alternative splicing patterns. Nucl Acids Res. 1991;19:4709–4716. doi: 10.1093/nar/19.17.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen OS. Neural cell adhesion molecule (NCAM) as a quantitative marker in synaptic remodeling. Neurochem Res. 1995;20:533–547. doi: 10.1007/BF01694535. [DOI] [PubMed] [Google Scholar]
- Kiss JZ, Muller D. Contribution of the neural cell adhesion molecule to neuronal and synaptic plasticity. Rev Neurosci. 2001;12:297–310. doi: 10.1515/revneuro.2001.12.4.297. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Mahtani MM, Nancarrow DJ, Brown DM, Kruglyak L, Mowry BJ, et al. Genome scan of schizophrenia. Am J Psychiatry. 1998;155:741–750. doi: 10.1176/ajp.155.6.741. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, et al. Genome scan meta-analysis of schizophrenia and bipolar disorder, Part II: Schizophrenia. Am J Hum Genet. 2003;73:34–48. doi: 10.1086/376549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziade M, Raymond V, Cliche D, Fournier JR, Caron C, Garneau Y, et al. Linkage results on 11q 21–22 in Eastern Quebec pedigrees densely affected by schizophrenia. Am J Med Genet. 1995;60:522–528. doi: 10.1002/ajmg.1320600607. [DOI] [PubMed] [Google Scholar]
- Moran N, Bock E. Characterization of the kinetics of neural cell adhesion molecule homophilic binding. FEBS Lett. 1988;242:121–124. doi: 10.1016/0014-5793(88)80998-0. [DOI] [PubMed] [Google Scholar]
- Muller D, Wang C, Skibo G, Toni N, Cremer H, Calaora V, et al. PSA-NCAM is required for activity-induced synaptic plasticity. Neuron. 1996;17:413–422. doi: 10.1016/s0896-6273(00)80174-9. [DOI] [PubMed] [Google Scholar]
- Muller D, Djebbara-Hannas Z, Jourdain P, Vutskits L, Durbec P, Rougon G, et al. Brain-derived neurotrophic factor restores long-term potentiation in polysialic acid-neural cell adhesion molecule-deficient hippocampus. Proc Natl Acad Sci USA. 2000;97:4315–4320. doi: 10.1073/pnas.070022697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ní Dhúill CM, Fox GB, Pittock SJ, O'Connell AW, Murphy KJ, Regan CM. Polysialylated neural cell adhesion molecule expression in the dentate gyrus of the human hippocampal formation from infancy to old age. J Neurosci Res. 1999;55:99–106. doi: 10.1002/(SICI)1097-4547(19990101)55:1<99::AID-JNR11>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Delling M, Sytnyk V, Dityatev A, Fukami K, Schachner M. Cosignaling of NCAM via lipid rafts and the FGF receptor is required for neuritogenesis. J Cell Biol. 2002;157:521–532. doi: 10.1083/jcb.200109059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odumeru O, Murphy KJ, O'Connell AW, Regan CM, Shiotani T. Influence of nefiracetam on NGF-induced neuritogenesis and neural cell adhesion molecule polysialic acid expression: in vivo and in vitro comparisons. Behav Brain Res. 1997;83:173–178. doi: 10.1016/s0166-4328(97)86064-0. [DOI] [PubMed] [Google Scholar]
- Pillai-Nair N, Panicker AK, Rodriguiz RM, Gilmore KL, Demyanenko GP, Huang JZ, et al. Neural cell adhesion molecule-secreting transgenic mice display abnormalities in GABAergic interneurons and alterations in behavior. J Neurosci. 2005;25:4659–4671. doi: 10.1523/JNEUROSCI.0565-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak M, Frye MA, Wright R, Hemperly JJ, George MS, Pazzaglia PJ, et al. Increased neural cell adhesion molecule in the CSF of patients with mood disorder. J Neurochem. 1996;66:1532–1538. doi: 10.1046/j.1471-4159.1996.66041532.x. [DOI] [PubMed] [Google Scholar]
- Prediger EA, Hoffman S, Edelman GM, Cunningham BA. Four exons encode a 93-base-pair insert in three neural cell adhesion molecule mRNAs specific for chicken heart and skeletal muscle. Proc Natl Acad Sci USA. 1988;85:9616–9620. doi: 10.1073/pnas.85.24.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes AA, Schulte SV, Small S, Akeson R. Distinct NCAM splicing events are differentially regulated during rat brain development. Brain Res Mol Brain Res. 1993;17:201–211. doi: 10.1016/0169-328x(93)90003-8. [DOI] [PubMed] [Google Scholar]
- Ronn LC, Pedersen N, Jahnsen H, Berezin V, Bock E. Brain plasticity and the neural cell adhesion molecule (NCAM). Adv Exp Med Biol. 1997;429:305–322. doi: 10.1007/978-1-4757-9551-6_22. [DOI] [PubMed] [Google Scholar]
- Ronn LC, Hartz BP, Bock E. The neural cell adhesion molecule (NCAM) in development and plasticity of the nervous system. Exp Gerontol. 1998;33:853–864. doi: 10.1016/s0531-5565(98)00040-0. [DOI] [PubMed] [Google Scholar]
- Rundus VR, Marshall GB, Parker SB, Bales ES, Hertzberg EL, Minkoff R. Association of cell and substrate adhesion molecules with connexin43 during intramembranous bone formation. Histochem J. 1998;30:879–896. doi: 10.1023/a:1003449525619. [DOI] [PubMed] [Google Scholar]
- Saffell JL, Williams EJ, Mason IJ, Walsh FS, Doherty P. Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron. 1997;18:231–242. doi: 10.1016/s0896-6273(00)80264-0. [DOI] [PubMed] [Google Scholar]
- Saito S, Tanio Y, Tachibana I, Hayashi S, Kishimoto T, Kawase I. Complementary DNA sequence encoding the major neural cell adhesion molecule isoform in a human small cell lung cancer cell line. Lung Cancer. 1994;10:307–318. doi: 10.1016/0169-5002(94)90660-2. [DOI] [PubMed] [Google Scholar]
- Sasieni P. From genotypes to genes: doubling the sample size. Biometrics. 1997;53:1253–1261. [PubMed] [Google Scholar]
- Seki T, Arai Y. Age-related production of new granule cells in the adult dentate gyrus. Neuroreport. 1995;6:2479–2482. doi: 10.1097/00001756-199512150-00010. [DOI] [PubMed] [Google Scholar]
- Stork O, Welzl H, Cremer H, Schachner M. Increased intermale aggression and neuroendocrine response in mice deficient for the neural cell adhesion molecule (NCAM). Eur J Neurosci. 1997;9:1117–1125. doi: 10.1111/j.1460-9568.1997.tb01464.x. [DOI] [PubMed] [Google Scholar]
- Stork O, Welzl H, Wotjak CT, Hoyer D, Delling M, Cremer H, et al. Anxiety and increased 5-HT1A receptor response in NCAM null mutant mice. J Neurobiol. 1999;40:343–355. doi: 10.1002/(sici)1097-4695(19990905)40:3<343::aid-neu6>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Terwilliger JD, Ott J. Handbook of human genetic linkage. Johns Hopkins University Press; Baltimore: 1994. pp. 188–193. [Google Scholar]
- Tomita H, Vawter MP, Walsh DM, Evans SJ, Choudary PV, Li J, et al. Effect of agonal and postmortem factors on gene expression profile: quality control in microarray analyses of postmortem human brain. Biol Psychiatry. 2004;55:346–352. doi: 10.1016/j.biopsych.2003.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duijnhoven HL, Helfrich W, de Leij L, Roebroek AJ, Van de Ven WJ, Healey K. Splicing of the VASE exon of neural cell adhesion molecule (NCAM) in human small-cell lung carcinoma (SCLC). Int J Cancer. 1992;50:118–123. doi: 10.1002/ijc.2910500124. [DOI] [PubMed] [Google Scholar]
- Vawter MP. Dysregulation of the neural cell adhesion molecule and neuropsychiatric disorders. Eur J Pharmacol. 2000;405:385–395. doi: 10.1016/s0014-2999(00)00568-9. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Hemperly JJ, Hyde TM, Bachus SE, VanderPutten DM, Howard AL, et al. VASE-containing N-CAM isoforms are increased in the hippocampus in bipolar disorder but not schizophrenia. Exp Neurol. 1998;154:1–11. doi: 10.1006/exnr.1998.6889. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Howard AL, Hyde TM, Kleinman JE, Freed WJ. Alterations of hippocampal secreted N-CAM in bipolar disorder and synaptophysin in schizophrenia. Mol Psychiatry. 1999;4:467–475. doi: 10.1038/sj.mp.4000547. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Mittal B, Walsh FS, Doherty P. FGF inhibits neurite outgrowth over monolayers of astrocytes and fibroblasts expressing transfected cell adhesion molecules. J Cell Sci. 1995;108:3523–3530. doi: 10.1242/jcs.108.11.3523. [DOI] [PubMed] [Google Scholar]
- Wittke-Thompson JK, Pluzhnikov A, Cox NJ. Rational inferences about departures from Hardy–Weinberg equilibrium. Am J Hum Genet. 2005;76:967–986. doi: 10.1086/430507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood GK, Tomasiewicz H, Rutishauser U, Magnuson T, Quirion R, Rochford J, et al. NCAM-180 knockout mice display increased lateral ventricle size and reduced prepulse inhibition of startle. Neuroreport. 1998;9:461–466. doi: 10.1097/00001756-199802160-00019. [DOI] [PubMed] [Google Scholar]
- Zacharias U, Norenberg U, Rathjen FG. Functional interactions of the immunoglobulin superfamily member F11 are differentially regulated by the extracellular matrix proteins tenascin-R and tenascin-C. J Biol Chem. 1999;274:24357–24365. doi: 10.1074/jbc.274.34.24357. [DOI] [PubMed] [Google Scholar]
- Zhao JH, Curtis D, Sham PC. Model-free analysis and permutation tests for allelic associations. Hum Hered. 2000;50:133–139. doi: 10.1159/000022901. [DOI] [PubMed] [Google Scholar]