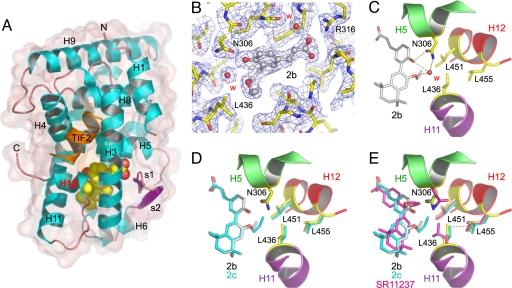
Fig. 4.
Structures of RXRα LBD in complex with partial agonists. (A) Overall structure of RXRα LBD in complex with 2a, 2b, or 2c. The ligand is represented by red (oxygen atoms) and yellow (carbon atoms) van der Waals spheres. Helices and β-strands are numbered from N to C terminus. Together, helices H3, H4, and H12 define the activation function 2 (AF-2) surface to which the TIF2 NR2 peptide is bound. (B) 2Fo−Fc density (1σ) for the LBP of RXRα bound to 2b. W indicates a water molecule. (C) Closeup view showing the hydrogen bond network that stabilizes a water molecule in close proximity of L436. An identical hydrogen bond network is observed in the complex with 2a (not shown). (D) Superposition of the RXR LBP with 2b and 2c. (E) Comparison with the structure of RXRα LBD bound to SR11237 (Protein Data Bank ID code 1MVC). To accommodate the particular features of 2a–c, L436 must adopt a conformation that differs from that found in the presence of the agonist. The dashed line between L436 and L455 indicates a short distance.