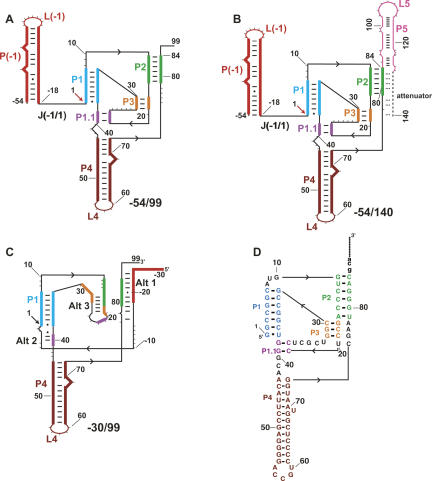
FIGURE 1.
Secondary structures present in HDV ribozyme-containing transcripts. The structures are color coded by native pairings, which are based on the crystal structure of the cleaved form of the ribozyme (Ferre-D'Amare et al. 1998) and extensive mutagenesis experiments (Wadkins et al. 1999; Chadalavada et al. 2000, 2002; Diegelman-Parente and Bevilacqua 2002; Brown et al. 2004). Black arrowheads denote 5′ to 3′ directionality. (A,B) Native secondary structures of constructs studied. These transcripts begin at −54, which allows formation of the P(−1) pairing that facilitates native secondary structure (Chadalavada et al. 2000). P(−1) has the L(−1) hairpin loop at its apex and is joined to the ribozyme by the J(−1/1) single-stranded joining region. Pairings P1, P2, P3, P4, and P1.1 are contained within the ribozyme, and the site of cleavage between −1 and 1 is denoted with an arrow. For the transcript in B, increasing length of downstream sequence is present, which results in formation of P5 and the attenuator. Pairing register of the attenuator, which is mutually exclusive with much of the ribozyme secondary structure, is denoted by an extension (dashed line). (C) Secondary structure of the −30/99 RNA showing some of the known alternative pairings. Alt 1 and Alt 2 involve interactions of upstream flanking sequence with the ribozyme, while Alt 3 involves ribozyme–ribozyme pairing (Chadalavada et al. 2000, 2002). Other alternative pairings not shown include Alt X and Alt Y, which disfavor native RNA folding (Brown et al. 2004). (D) Sequence of wild-type ribozyme (Chadalavada et al. 2002).