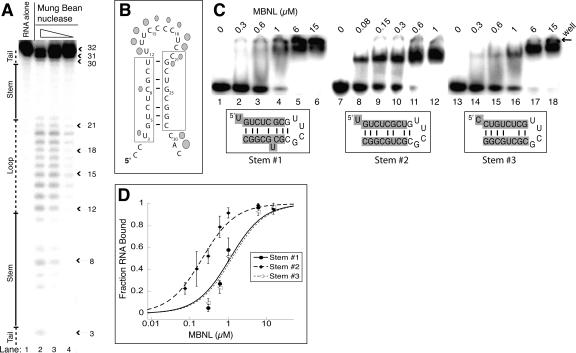
FIGURE 5.
MBNL binds the cTNT 32mer as a stem–loop. (A) Mung bean nuclease cleavage pattern of cTNT 32mer. The concentration of mung bean nuclease is 100, 10, and 1 units per microliter, in lanes 2–4, respectively. (B) Schematic of the likely stem–loop within cTNT intron 4, showing cleavage locations of mung bean nuclease. Larger shapes indicate strong cleavage events while smaller shapes represent weak cleavage events. The cross-linking sites determined previously are boxed (Ho et al. 2004). (C) Binding curve of the three potential stem structures of the cTNT intron 4, showing MBNL prefers structure #2. (D) Gel shifts showing MBNL binding to the three potential stems of cTNT intron 4. Concentration of MBNL is labeled above each lane; note the lower concentrations in lanes 8–12 compared to lanes 2–6 and lanes 14–18. Sequences highlighted in gray are sequences from the cTNT intron in the three different potential base pair and mismatch configurations. The nongray sequence is the tetraloop cap and an additional base pair to stabilize the structure if necessary.