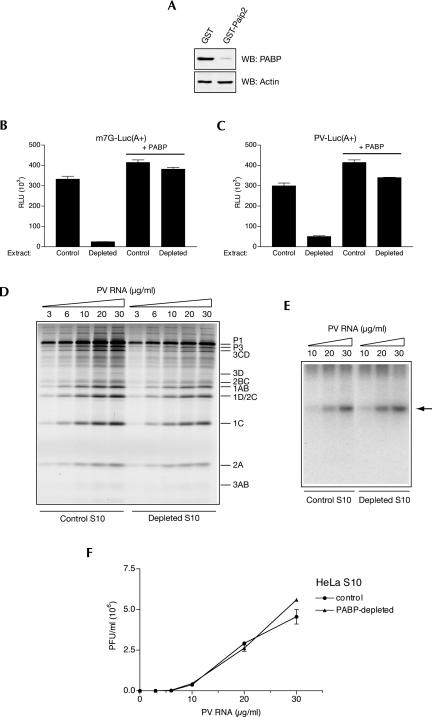
FIGURE 4.
Effect of PABP depletion on PV genome expression in nuclease-treated HeLa S10 extract. (A) Extracts were treated with GST or GST-Paip2 coupled to beads and their aliquots (4 μL, 50 μg of protein) were analyzed by Western blotting for PABP or actin as indicated. The degree of PABP depletion was 96%. (B) Cap-dependent translation of a reporter mRNA is strongly reduced by PABP depletion. Capped poly(A+) luciferase mRNA (1 μg/mL) was translated in control or PABP-depleted extract in the absence or presence of PABP as indicated. Translation conditions and luciferase assay are described in Fig. 3B. (C) PV IRES-mediated translation of a reporter mRNA is significantly reduced by PABP depletion. Control and PABP-depleted extracts were programmed with PV IRES-containing poly(A+) luciferase mRNA (1 μg/mL) in the absence or presence of PABP as indicated. For translation conditions and luciferase assay, see Fig. 3C. (D) PV RNA translation in PABP-depleted extracts. Control and PABP-depleted S10 extracts were programmed with the indicated concentrations of PV RNA in the presence of [35S]methionine. Labeled PV proteins were analyzed by SDS-PAGE and fluorography; their positions are indicated to the right of the gel. Endogenous mRNA translation yielded no products (data not shown). (E) RNA synthesis in control and PABP-depleted extracts programmed with the indicated concentrations of PV RNA. [α-32P]CTP pulse labeling of RNA was between 4 and 5 h of incubation. Product RNA analysis was as for Fig. 2B. (Arrow) Position of single-stranded PV RNA. (F) PV RNA-directed PV synthesis is not diminished by PABP depletion. Control and PABP-depleted extracts were programmed with the indicated concentrations of PV RNA. PV titers were determined in triplicate using HeLa R19 cell monolayers. Averages of the obtained values (with standard deviation from the mean) are shown.