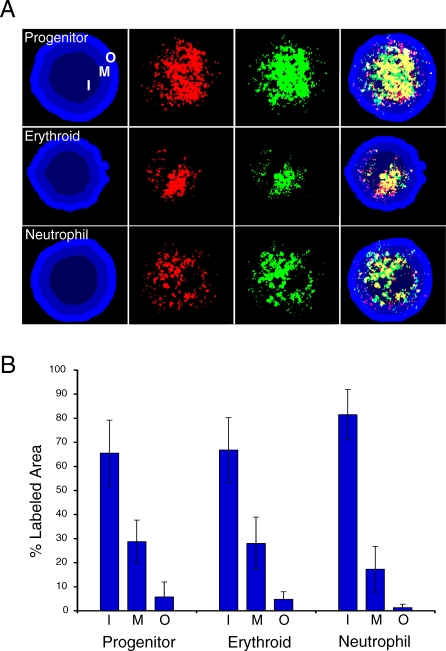
Figure 3. Gene Expression Is Concentrated in the Nuclear Center.
We analyzed the organization of total gene activity in the progenitor, erythroid, and neutrophil cell types through a modified FISH approach (Materials and Methods).
(A) Progenitor, erythroid, and neutrophil nuclei hybridized with biotin- (green) and digoxigenin-conjugated (red) probe material generated from cDNA isolated from each cell type. Cohybridizing with differently labeled probes controls for the specificity of detection (i.e., yellow indicates the degree of overlap for the two probes). The nucleus is counterstained with 4-,6-diamidino-2-phenylindole (DAPI), and the nucleus is divided into three areas of equal area, indicated as inner (I), middle (M), and outer (O). An algorithm in SVCell was created to make these shells and determine the percentage of pixels of the probe material that is localized to the three nuclear regions.
(B) Bar graph of results expressed as the percentage of total area in each region. At least 30 nuclei were analyzed per lineage; lines indicate standard deviation.