Abstract
Plants sense the presence of potentially competing nearby individuals as a reduction in the red to far-red ratio of the incoming light. In anticipation of eventual shading, a set of plant responses known as the shade avoidance syndrome (SAS) is initiated soon after detection of this signal by the phytochrome photoreceptors. Here we analyze the function of PHYTOCHROME RAPIDLY REGULATED1 (PAR1) and PAR2, two Arabidopsis thaliana genes rapidly upregulated after simulated shade perception. These genes encode two closely related atypical basic helix–loop–helix proteins with no previously assigned function in plant development. Using reverse genetic approaches, we show that PAR1 and PAR2 act in the nucleus to broadly control plant development, acting as negative regulators of a variety of SAS responses, including seedling elongation and photosynthetic pigment accumulation. Molecularly, PAR1 and PAR2 act as direct transcriptional repressors of two auxin-responsive genes, SMALL AUXIN UPREGULATED15 (SAUR15) and SAUR68. Additional results support that PAR1 and PAR2 function in integrating shade and hormone transcriptional networks, rapidly connecting phytochrome-sensed light changes with auxin responsiveness.
Keywords: Arabidopsis , auxin, phytochrome, SAUR , shade avoidance
Introduction
The shade avoidance syndrome (SAS) refers to a set of plant responses aimed to adapt growth and development to environments of high plant density, like those found in both natural (e.g., forests) and agricultural (e.g., orchards) communities. Presence of nearby plants results in a reduction in the red (R) to far-red (FR) ratio (R:FR) caused by a specific enrichment in FR light reflected from the surface of neighboring leaves. The R:FR changes are perceived by the phytochrome photoreceptors (Smith, 1982; Smith and Whitelam, 1997). The phytochromes detect the R and FR parts of the spectrum and have a major role in controlling adaptive responses such as seed germination, stem elongation, leaf expansion and flowering time. In Arabidopsis (Arabidopsis thaliana), a small gene family of five members encodes the phytochromes (PHYA-PHYE) (Quail, 2002; Chen et al, 2004). Although phyB is the major phytochrome controlling SAS, genetic and physiological analyses have shown that other phytochromes act redundantly with phyB in the control of some aspects of SAS-driven development, like flowering time (phyD, phyE), petiole elongation (phyD, phyE) and internode elongation between rosette leaves (phyE) (Smith and Whitelam 1997; Devlin et al, 1998, 1999).
Downstream of R:FR perception by phytochromes, expression of several genes has been shown to rapidly and reversibly change in response to simulated shade (Devlin et al, 2003; Salter et al, 2003). Although it is unclear as to what extent the changes in the expression of these PHYTOCHROME RAPIDLY REGULATED (PAR) genes are instrumental for implementing the morphological and physiological SAS responses, it is likely that these photoresponses are a consequence of the regulation of a complex transcriptional network by phytochromes, as postulated for seedling de-etiolation (Quail, 2002; Jiao et al, 2007). Indeed, genetic approaches have demonstrated roles in SAS for some PAR genes encoding transcription factors, including ATHB2, HFR1 and PIL1. A positive role for ATHB2 has been proposed based on overexpression studies (Steindler et al, 1999), whereas a negative role for HFR1 and PIL1 was deduced based on the elongation responses of mutant hypocotyls to shade (Sessa et al, 2005; Roig-Villanova et al, 2006). The low R:FR perception might therefore rapidly change the balance of positive and negative factors, resulting in the appropriate SAS responses. Genetic analyses have recently identified CSA1 as an SAS component that regulates the expression of ATHB2 and HFR1 (Faigón-Soverna et al, 2006), participating by unknown mechanisms in the shade-modulated transcriptional network. Eventually, this transcriptional network intersects with those of the major plant hormones, which regulate cell division and expansion changes needed for the specific photoresponses, that is, stem elongation and/or other changes to overgrow nearby competition. Indeed, several studies have established multiple links that connect auxin, brassinosteroid (BR), ethylene and gibberellins in the regulation of different photomorphogenic responses and specifically in the SAS responses (Tian and Reed, 2001; Devlin et al, 2003; Halliday and Fankhauser, 2003). Nonetheless, the precise molecular links for the interaction between SAS and hormonal transcriptional networks are mostly unknown.
Based on de-etiolation studies, it has been postulated that nuclear-localized phytochromes can potentially access G-box regulatory motifs located in the promoter regions of master regulatory genes by directly interacting with phytochrome-interacting factors (PIFs) (Martínez-García et al, 2000). One of them, PIF3, was the founder of a subgroup of basic helix–loop–helix (bHLH) proteins that act as regulators of seedling de-etiolation (Ni et al, 1998; Fairchild et al, 2000; Huq and Quail, 2002; Kim et al, 2003; Salter et al, 2003; Huq et al, 2004). This subfamily, known as group VII or subfamily 15 (Heim et al, 2003; Toledo-Ortiz et al, 2003), includes PIFs (i.e., PIF1, PIF4) and PIF3-like proteins (PILs), which lack the ability to directly interact with the phytochromes (i.e., HFR1, PIL1), but have been shown or proposed to heterodimerize with PIFs, potentially modulating the bHLH network activity. All the known bHLHs (PIFs and PILs) involved in phytochrome-mediated light signaling belong to this subfamily. The bHLH domain that defines the bHLH class of transcription factors encompasses ca. 60 amino acids arranged in two subdomains: a basic N-terminal stretch of 15–20 residues and an HLH domain composed of two amphipathic alpha helices separated by a variable loop region. The basic domain is involved in DNA binding, whereas the HLH domain is required for protein–protein interaction (i.e., dimerization). Comparison of animal bHLH sequences led to a hypothetical consensus motif of 19 conserved residues that define the bHLH domain (Atchley et al, 1999). Different groups of plant bHLHs fit this consensus (Buck and Atchley, 2003; Heim et al, 2003; Toledo-Ortiz et al, 2003). Several subfamilies of Arabidopsis bHLH proteins show conserved motifs outside the bHLH domain that might provide additional DNA-binding ability and specificity and/or protein interaction activities (Heim et al, 2003; Toledo-Ortiz et al, 2003). For instance, several members of the Arabidopsis bHLH subfamily 15 have an active phytochrome-binding motif shown to be necessary for PIF4 function in phyB signaling (Khanna et al, 2004).
We previously identified PAR1, a direct target gene of phytochrome action, whose expression is rapidly upregulated by shade (Roig-Villanova et al, 2006). In this work we show that PAR1 and its close relative PAR2 are atypical bHLH proteins that negatively control growth and metabolic SAS responses. In addition, we found that they act in the nucleus, impairing the auxin-regulated expression of two SMALL AUXIN UPREGULATED (SAUR) genes.
Results
PAR1 and PAR2 are novel bHLH-like proteins
The PAR1 gene (At2g42870), previously identified to be a primary target of phytochrome signaling (Roig-Villanova et al, 2006), encodes a short protein of 118 residues of unknown function. PAR1 is closely related in sequence to another Arabidopsis gene that we have named PAR2 (At3g58850), which encodes a protein of the same size and with 72% similarity (64% identity) to PAR1 (Supplementary Figure S1). Based on the existence of ESTs and our own data (www.arabidopsis.org; Roig-Villanova et al, 2006), we concluded that both PAR1 and PAR2 are expressed genes with no introns and short 5′ and 3′ UTRs. As shown in Figure 1A, the expression of PAR1 and PAR2 was rapidly upregulated after a simulated shade treatment of FR-enriched white light (W+FR), consistent with their classification as PAR genes (Roig-Villanova et al, 2006). PAR2 upregulation, however, was slower and weaker compared to that of PAR1 (Figure 1A).
Figure 1.
Expression and sequence analyses of PAR1 and PAR2. (A) RNA blot analysis of PAR1 and PAR2 expression in 7-day-old W-grown wild-type seedlings treated with W+FR for 0, 1, 2 and 3 h. 25S rRNA levels are shown as a loading control. (B) Multiple sequence alignment of bHLH domains from Arabidopsis group VIII proteins (Heim et al, 2003), PAR1 and PAR2. Identical residues are boxed in black. Gray boxes mark partially conserved residues. The position of the basic, helix and loop regions is indicated. (C) Neighbor joining unrooted phylogenetic tree based on the alignment shown in Supplementary Figure S2. Only the monophyletic clade grouping PAR1, PAR2 and group VIII bHLHs is shown. Branch lengths are not proportional to the distance between sequences. Numbers in nodes correspond to bootstrap support values indicated as percentages.
As a first step to investigate the functional identity of PAR1 and PAR2 proteins, InterProScan searches (www.ebi.ac.uk/InterProScan/) were performed, identifying a region corresponding to a bHLH domain (IPR011598). When PAR1 and PAR2 were used as queries in PSI-BLAST searches, only sequence hits corresponding to Arabidopsis proteins of the bHLH group were retrieved with significant scores. PAR1 and PAR2 are most similar to these transcription factors within the HLH region (Figure 1B; Supplementary Figure S2A), sharing most of the conserved sites and fitting well to the hypothetical, predictive consensus motif previously proposed (Atchley et al, 1999; Toledo-Ortiz et al, 2003). By contrast, PAR1 and PAR2 sequences diverged in the basic region, lacking the H/K9-E13-R17 motif characterized as critical for proper DNA binding (Ferre-D'Amare et al, 1993; Atchley et al, 1999; Toledo-Ortiz et al, 2003).
The predicted bHLH-like domains of PAR1, PAR2 and 133 putative Arabidopsis bHLHs were aligned (Supplementary Figure S2), generating a phylogenetic tree with a similar topology to that previously reported (Heim et al, 2003). As shown in Figure 1C, PAR1 and PAR2 are included within subgroup VIII-A (Heim et al, 2003), which corresponds to subfamilies 19 and 20 from a different classification (Toledo-Ortiz et al, 2003). No function has been proposed for any of the members of this subgroup. An MEME analysis (http://bioweb.pasteur.fr/seqanal/motif/meme) to search for highly conserved regions outside the bHLH-like domain within group VIII showed no conserved motifs (Supplementary Figure S3).
Overexpression of PAR1 and PAR2 results in dwarf dark-green plants
To investigate the role of PAR1 in planta, we generated plants constitutively overexpressing PAR1 alone, or as an N-terminal fusion with the green fluorescent protein (GFP) and the GFP-GUS double reporter (P35S:PAR1, P35S:PAR1-G and P35S:PAR1-GG lines, respectively) (Figure 2). Transgenic plants presented a characteristic dwarf phenotype with compact rosettes and inflorescences, epinastic leaves, shorter flowering stems and siliques and a general dark-green color (Figure 2). This phenotype was most frequent when the untagged PAR1 protein was used (data not shown). The most severely affected lines grew slowly and their short siliques resulted in reduced seed production, complicating the isolation of homozygous lines. All the lines overexpressing the transgene displayed a dwarf dark-green phenotype, supporting that these traits were caused by PAR1 overexpression. Similar results were obtained with P35S:PAR2 and P35S:PAR2-G plants overexpressing PAR2 alone, or as an N-terminal fusion with GFP (Figure 2). These data suggest that both PAR1 and PAR2 might play a similar role in plant development.
Figure 2.
Phenotype of plants overexpressing PAR1 or PAR2. (A) Molecular characterization of PAR1 and PAR2 overexpressing plants. RNA extracted from 7-day-old W-grown wild-type (wt) or transgenic seedlings was used for RNA blot analysis of PAR1 and PAR2 expression levels. Each RNA sample was extracted from a pool of seedlings corresponding to a segregating population. 25S rRNA levels are shown as a loading control. (B) Representative 7-day-old W-grown wt and transgenic seedlings overexpressing the indicated version of PAR1 or PAR2. (C) Adult wt and transgenic plants overexpressing the indicated version of PAR1 or PAR2 grown for 6 (upper panel) or 8 weeks (lower panel) under short-day conditions. In each section panels are to the same scale.
Phenotypic traits are oppositely affected by PAR1 or PAR2 overexpression and simulated shade
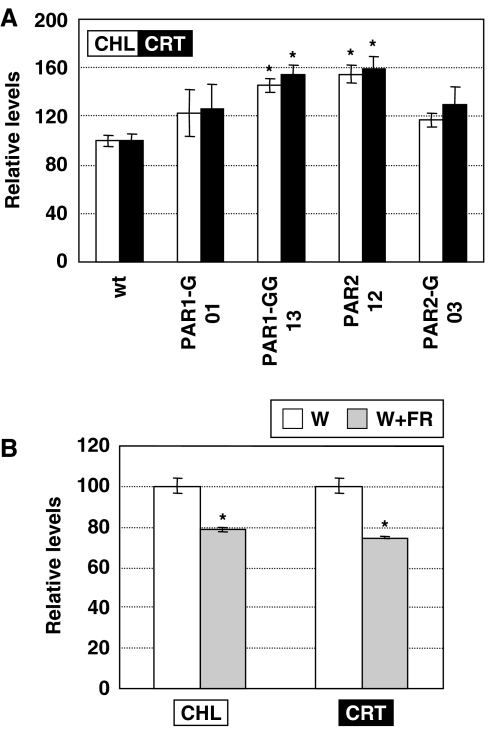
The most obvious phenotypes of PAR1-overexpressing seedlings grown under continuous white light (W) were a short hypocotyl length and a strongly reduced cotyledon and primary leaf longitudinal expansion (Figures 2B and 3), which ultimately resulted in the dwarf phenotype observed. The dark-green phenotype of these lines suggested that the levels of photosynthetic pigments might also be altered in transgenic plants. As predicted, measurements of chlorophyll and carotenoid levels in W-grown seedlings demonstrated that transgenic seedlings accumulated more chlorophylls and carotenoids than wild-type plants (Figure 4A). All these phenotypes were typically enhanced in the lines overexpressing PAR2 (Figures 3 and 4).
Figure 3.
Morphological phenotype of seedlings overexpressing PAR1 or PAR2. wt and transgenic seedlings were germinated and grown for 2 days under W and then either kept in W (white bars) or transferred to W+FR (gray bars) for 5 more days. At least 15 seedlings for each treatment were used to measure the length of their hypocotyls (Hyp), cotyledons (Cot) and primary leaves (PL). Columns represent the mean and bars represent twice the standard error of the mean (2 × s.e.) of the data. Asterisks indicate highly significant differences (P<0.01) relative to the corresponding wt controls.
Figure 4.
Photosynthetic pigment levels in seedlings overexpressing PAR1 or PAR2. (A) Chlorophyll (CHL) and carotenoid (CRT) levels of 7-day-old W-grown wt and transgenic seedlings. (B) CHL and CRT levels of wt seedlings grown as described in Figure 3. Values are means and s.e. of three (A) and four (B) independent samples. Values for W-grown wt seedlings were taken as 100. Asterisks indicate significant differences (P<0.05) relative to the corresponding controls.
The observed effects of PAR1 and PAR2 overexpression on these aspects of Arabidopsis development and metabolism led us to analyze whether these traits were associated with the response of wild-type plants to simulated shade. We observed that besides promoting hypocotyl elongation, simulated shade clearly induced cotyledon and primary leaf longitudinal expansion in wild-type seedlings (Figure 3). Chlorophyll accumulation has also been reported to be affected by shade (Smith and Whitelam, 1997). We noticed that Arabidopsis seedlings became paler after prolonged exposure to W+FR. Consistently, a significant decrease in the amount of chlorophylls and carotenoids was observed in wild-type seedlings after simulated shade treatment compared to W-grown controls (Figure 4B). These data indicate that simulated shade and PAR1 or PAR2 overexpression have opposite effects in SAS-related responses in Arabidopsis seedlings. Furthermore, the response of transgenic seedlings to W+FR in terms of hypocotyl, cotyledon and primary leaf elongation was clearly attenuated compared to the wild type (Figure 3), suggesting that PAR1 and PAR2 may act as negative regulators of SAS.
Decreased PAR1 and PAR2 transcript levels result in enhanced SAS responses
To analyze the consequence of reduced PAR1 and PAR2 levels in Arabidopsis, we initially targeted the PAR1 gene for silencing, using an RNAi approach. Seven PAR1-RNAi lines with a single T-DNA insertion were generated. To select for silenced lines among them, PAR1 transcript levels were evaluated by RNA blot analyses in seedlings treated with W+FR for 1 h. The level of endogenous PAR1 transcripts was mildly reduced in three PAR1-RNAi lines tested compared to the wild type (Figure 5A). Two extra bands recognized by the PAR1 probe were also observed in some lines (Figure 5A), likely corresponding to the expression of the PAR1-RNAi construct. Analysis of PAR2 expression in the same lines also showed reduced transcript levels (Figure 5A), indicating that the PAR1-RNAi construct partially silenced both PAR1 and PAR2 genes. The most apparent phenotype of adult PAR1-RNAi plants grown under our greenhouse conditions was reduced fertility, that is, seed production (Figure 5C), and, as described for sterile mutants (Hensel et al, 1994), increased branching (data not shown). In seedlings, reduced PAR1 and PAR2 levels resulted in only slightly longer hypocotyls compared to wild-type controls (Figure 5B). Cotyledon and primary leaf longitudinal expansion phenotypes, however, were more strongly affected in PAR1-RNAi lines, as shown for overexpression lines. Under W, transgenic seedlings had longer cotyledons and primary leaves than the wild type, a phenotype similar to that induced by simulated shade. These differences were clearly increased under W+FR (Figure 5B), supporting the fact that reduced PAR1 and PAR2 transcript levels resemble an enhanced response to W+FR treatment.
Figure 5.
Characterization of PAR1-RNAi lines. (A) RNA blot analysis of PAR1 and PAR2 expression in 7-day-old W-grown seedlings treated with W+FR for 1 h. 25S rRNA levels are shown as a loading control. (B) Length of hypocotyls (Hyp), cotyledons (Cot) and primary leaves (PL) of wt and independent PAR1-RNAi lines grown as indicated in Figure 3. Mean and 2 × s.e. of at least 15 seedlings for each treatment are shown. (C) Seed production of wt and independent PAR1-RNAi lines grown under greenhouse conditions. Mean and s.e. of at least seven plants for each line are shown. Asterisks indicate significant (*P<0.05) or highly significant (**P<0.01) differences relative to the corresponding wt plants.
In the course of this study, we found a SALK line with a T-DNA insertion in the promoter region of PAR2, about 350 nucleotides upstream of the ATG codon (Figure 6A). Northern blot analysis showed no detectable PAR2 expression, whereas PAR1 transcript levels were unaffected (Figure 6B). These results indicate that this line, which we named par2-1, is probably a null mutant for PAR2. Adult par2-1 plants grown in the greenhouse showed no reduction in fertility (data not shown), suggesting that this trait is either redundantly regulated by both PAR1 and PAR2, or is due to off-target effects of the PAR1-RNAi construct. Under W, par2-1 seedlings showed slightly but significantly longer hypocotyls, cotyledons and primary leaves compared with wild-type controls. Also, as shown for PAR1-RNAi lines, these differences were clearly increased under W+FR (Figure 6C), arguing in favor of a specific negative role of PAR1 and PAR2 in the SAS-related traits analyzed. Two-way ANOVA tests indicated a significant (P<0.05) interaction between low levels of PAR1 and/or PAR2 and simulated shade treatments (Figures 5B and 6C) in the case of cotyledon and primary leaf elongation responses. Hypocotyl elongation under W+FR was significantly different to that under W in par2-1 seedlings, but not in PAR1-RNAi lines, when compared with the wild type. This statistical analysis also confirmed a highly significant interaction between increased levels of PAR1 or PAR2 and light treatments in all the three traits analyzed (Figure 3). Together, our data show that altered levels of PAR1 and/or PAR2 significantly affect seedling responsiveness to simulated shade.
Figure 6.
Characterization of par2 and hfr1 mutant seedlings. (A) Schematic representation of PAR2 (At3g58850) genomic sequence and T-DNA insertion in par2-1. The intronless coding sequence (light-gray box) and the position of oligonucleotides used for genotyping are shown. (B) RNA blot analysis of PAR1 and PAR2 expression in 7-day-old W-grown wt and par2-1 seedlings after W+FR treatment for 4 h. 25S rRNA levels are shown as a loading control. (C, D) Length of hypocotyls (Hyp), cotyledons (Cot) and primary leaves (PL) of mutant par2-1 (C) and hfr1 seedlings (D) grown as indicated in Figure 3. Mean and 2 × s.e. of at least 15 seedlings for each treatment are shown. Asterisks indicate significant (*P<0.05) or highly significant (**P<0.01) differences relative to the corresponding wt plants.
To compare the phenotypic effect of reduced PAR1 and PAR2 levels with that of known negative regulators of SAS, we analyzed hypocotyl, cotyledon and primary leaf length in hfr1-4 and hfr1-5 mutant seedlings grown under our specific experimental (light) conditions. Under W, the null hfr1-5 mutant (Sessa et al, 2005) showed slightly longer hypocotyls, cotyledons and primary leaves compared with wild-type controls, whereas seedlings of the hfr1-4 mutant (which shows low levels of HFR1 transcripts; Sessa et al, 2005) were indistinguishable from the wild type. Under W+FR, these traits were significantly enhanced only in hfr1-5 seedlings (Figure 6D). These data indicate that after simulated shade treatments, loss of HFR1 function results in SAS phenotypes qualitatively and quantitatively similar to those displayed by seedlings with reduced PAR1 and/or PAR2 expression. In summary, the results with transgenic lines in which PAR1 and PAR2 levels are altered support a role for these atypical bHLH proteins as negative regulators of SAS responses in Arabidopsis seedlings.
PAR1 and PAR2 proteins function in the nucleus
Because of the sequence similarity of PAR1 and PAR2 to members of the bHLH family of transcription factors, we hypothesized that these two proteins could regulate development by modulating gene transcription. We initially investigated whether PAR1 and PAR2 were nuclear proteins. Taking advantage of our transgenic lines overexpressing functional chimeras of PAR1 and PAR2 fused to GFP, we examined the subcellular localization of the fusion proteins in the roots of P35S:PAR1-G and P35S:PAR2-G plants with a dwarf phenotype. As controls, we used transgenic P35S:GFP and P35S:GUS-GFP plants. As expected, cytoplasmic and nuclear localization were observed for GFP, whereas the larger size of the GUS-GFP fusion prevented its diffusion into the nucleus (Figure 7A). Both PAR1-GFP and PAR2-GFP proteins were mainly localized in the nuclei (Figure 7A). Nuclear localization of the fusion proteins was also observed in shoot tissues (Supplementary Figure S4) and it was not affected by light conditions (data not shown).
Figure 7.
Subcellular localization and function of tagged versions of PAR1 and PAR2. (A) GFP fluorescence in roots from 7-day-old W-grown transgenic seedlings expressing the indicated GFP-tagged proteins. Panels are to the same scale. (B) Phenotype of 7-day-old W-grown wt and transgenic seedlings overexpressing PAR1-GR germinated and grown on medium either supplemented (+) or not (−) with 5 μM DEX.
To confirm whether nuclear localization was required for PAR1 function, a translational fusion between PAR1 and the glucocorticoid receptor (GR) domain was constitutively expressed in plants (P35S:PAR1-GR lines). The GR domain typically retains a nuclear factor in the cytoplasm in the absence of a steroid ligand, but nuclear localization is restored in the presence of the synthetic glucocorticoid dexamethasone (DEX). In the absence of DEX, most transgenic P35S:PAR1-GR seedlings were similar to wild-type seedlings (Figure 7B), suggesting that accumulation of cytosolic PAR1-GR did not have a visible effect on plant development or pigment accumulation. By contrast, DEX treatment only induced a severe dwarf dark-green phenotype in P35S:PAR1-GR seedlings (Figure 7B). From a total of 10 independent transgenic lines with a single T-DNA insertion isolated, five clearly showed a dwarf dark-green phenotype upon DEX treatment. In some lines we observed a mild phenotype in the absence of DEX, an effect attributed to the production of truncated, constitutively active versions of the fusion protein (Sablowski and Meyerowitz, 1998). In these lines, however, DEX treatment dramatically enhanced the transgenic phenotype (data not shown). Together, these results demonstrate that PAR1 is active only when localized in the nucleus, suggesting a role for both PAR1 and PAR2 as transcriptional regulators.
PAR1 and PAR2 repress hormone-mediated upregulation of SAUR genes
To look for potential targets of PAR1 action, we analyzed global transcript profiles in W-grown wild-type and dwarf P35S:PAR1-GG seedlings (a transgenic line already available at that time) to screen for differentially expressed genes. From the 120 genes identified as being misregulated in the P35S:PAR1-GG seedlings (Supplementary Table S1), 34 (28%) had been previously identified by other authors as regulated by auxin and/or BR (Nemhauser et al, 2004, 2006; Supplementary Table S2). Most of these (30 genes) belong to the subgroup of 70 genes downregulated in P35S:PAR1-GG seedlings; from these, 22 genes are upregulated by auxins and/or BR with almost half of them (10) upregulated by both the hormones (Supplementary Figure S5). Most strikingly, seven of these 10 genes belong to the SAUR family (McClure and Guilfoyle, 1987; Supplementary Table S2). For further experiments we focused on SAUR15 (also named SAUR-AC1; At4g38850) and SAUR68 (At1g29510), for which a 30–40% reduction in their expression level was detected in the microarray (Supplementary Table S1). Because the low expression levels of these genes did not allow us to conclusively validate changes between wild-type and PAR1-overexpressing seedlings by RNA blot analysis, we tested whether the auxin-dependent induction of these SAUR genes was affected in transgenic lines. As shown in Figure 8A, the upregulation of both SAUR15 and SAUR68 in response to treatment with 2,4-D (a synthetic auxin) was clearly attenuated when PAR1 or PAR2 was overexpressed. Similar results were observed after treatment with brassinolide (data not shown). By contrast, PAR1 or PAR2 overexpression did not affect the auxin-mediated upregulation of HAT2 (Figure 8A), another gene,which is rapidly induced by auxin (Sawa et al, 2002). Transcript levels for SAUR15 and SAUR68 were also lower in par2-1 line compared with the wild type, both before and after 2,4-D treatments (Figure 8B). Altogether, our results indicate that a certain level of PAR1 and/or PAR2 is required for the normal response of a subset of auxin-regulated genes (including SAUR15 and SAUR68 but not HAT2) to increased 2,4-D levels.
Figure 8.
Effect of PAR1, PAR2, auxin and simulated shade on SAUR expression. Seven-day-old W-grown seedlings were treated with either 50 μM 2,4-D (A, B) or W+FR (C, D) for the indicated times (in h). (A) RNA blot analysis of SAUR15, SAUR68 and HAT2 expression in auxin-treated wt and transgenic seedlings overexpressing PAR1-GG (line 13) and PAR2-G (line 03). (B) RNA blot analysis of SAUR15 and SAUR68 expression in auxin-treated wt and par2-1 seedlings. (C) RNA blot analysis of SAUR15 and SAUR68 expression in wt seedlings after simulated shade (W+FR). (D) RNA blot analysis of SAUR15 and SAUR68 expression in seedlings from the lines described in panel A treated with W+FR. 25S rRNA levels are shown as a loading control.
Interestingly, the expression of SAUR15 and SAUR68 is also rapidly but transiently induced by simulated shade (Figure 8C), which indicates that these two SAUR genes are also authentic PAR genes. The acute SAUR15 and SAUR68 expression response to simulated shade was clearly attenuated in PAR1- and PAR2-overexpressing lines (Figure 8D), which suggests that the observed PAR1- and PAR2-mediated downregulation of SAUR expression is meaningful for the regulation of SAS responses.
To address whether the observed negative role of PAR1 on SAUR gene expression was an early (direct) or late (indirect) effect on transcription, SAUR15 and SAUR68 transcript levels were monitored, following the targeting of PAR1-GR to the nucleus by DEX treatment of P35S:PAR1-GR seedlings. Transgenic and wild-type seedlings were first treated with 2,4-D to induce SAUR gene expression and 2 h later they were either treated or not with DEX (Figure 9A). In wild-type seedlings, auxin-induced expression of SAUR15, SAUR68 and HAT2 was unaffected by DEX application. By contrast, expression of SAUR15 and SAUR68 was reduced 4 h after DEX treatment in P35S:PAR1-GR seedlings (time point 6 h), whereas that of HAT2 was unaffected (Figure 9A). A similar experiment was performed in the presence of the protein synthesis inhibitor cycloheximide (CHX). Transgenic seedlings were first treated with 2,4-D to induce SAUR expression, and 2 h later they were either treated or not with DEX and/or CHX (Figure 9B). In the absence of CHX, addition of DEX only reduced the expression of SAUR15 and SAUR68, as expected. In the presence of CHX, expression of all three genes (SAUR15, SAUR68 and HAT2) was increased, as previously reported (Zimmermann et al, 2004). Addition of both DEX and CHX only repressed SAUR15 and SAUR68 expression (Figure 9B). We concluded that the DEX-dependent repression of SAUR15 and SAUR68 does not require de novo protein synthesis, consistent with these two SAUR genes being direct targets of PAR1 action.
Figure 9.
Identification of the primary targets of PAR1. (A) RNA blot analysis of the effect of DEX-dependent nuclear translocation of PAR1 on 2,4-D-induced expression of SAUR15, SAUR68 and HAT2 in 7-day-old W-grown wt and PAR1-GR overexpressing seedlings treated with 50 μM 2,4-D, incubated for 2 h and then either treated (+) or not (−) with 5 μM DEX. (B) RNA blot analysis of the effect of CHX on DEX-dependent repression of SAUR15 and SAUR68 in 7-day-old W-grown PAR1-GR overexpressing seedlings treated with 10 μM 2,4-D, incubated for 2 h and then either treated (+) or not (−) with 5 μM DEX in the absence (−CHX) or presence (+CHX) of 50 μM CHX. Plant material for RNA extraction was harvested at the time points indicated with asterisks. 25S rRNA levels are shown as a loading control.
Discussion
Despite the importance of SAS for plant survival, we know relatively little about the genetic components involved in its control. In contrast with the information available for other photomorphogenic responses, like seedling de-etiolation, genetic and molecular approaches have identified few SAS regulators, including ATHB2 (Steindler et al, 1999), HFR1 (Sessa et al, 2005) and PIL1 (Salter et al, 2003; Roig-Villanova et al, 2006). These proteins belong to two different families of transcription factors: homeodomain (ATHB2) and bHLH (HFR1 and PIL1). In this paper, we report the characterization of PAR1 and PAR2, two atypical bHLH-like proteins localized within the nucleus, with a role in the integration of light and auxin signaling.
The bHLH proteins represent one of the largest transcription factor families found in nature. They are widely distributed in all the eukaryotic kingdoms and control a great diversity of biological processes. In plants, the best characterized group is probably group VII (Heim et al, 2003) or subfamily 15 (Toledo-Ortiz et al, 2003), which includes HFR1, PIL1 and all the other PIFs and PILs involved in phytochrome signaling. PAR1 and PAR2 are classified here as part of group VIII-A (Figure 1C), whose members have a single exon encoding the bHLH domain. PAR1 and PAR2 are localized in regions of chromosomes 2 and 3, respectively, which have been subjected to segmental duplications (Blanc et al, 2000; Vision et al, 2000), suggesting that they are the result of a recent gene duplication event. To our knowledge, there is no previous functional information about the members of group VIII.
HFR1 and PIL1 act as negative regulators of SAS responses, whereas a positive role has been proposed for ATHB2 (Steindler et al, 1999; Sessa et al, 2005; Roig-Villanova et al, 2006). All these factors, as well as PAR1, were first identified based on their very rapid response to shade in terms of gene expression (Carabelli et al, 1996; Salter et al, 2003; Sessa et al, 2005; Roig-Villanova et al, 2006). All but HFR1 were demonstrated to be direct targets of phytochrome signaling (Roig-Villanova et al, 2006). PAR2, the closest PAR1 relative in the Arabidopsis genome, is also induced by shade, although more slowly than PAR1 (Figure 1). Alteration of PAR1 and PAR2 levels, however, similarly affects plant development, with a particularly clear effect on elongation and pigmentation responses. The traits affected by PAR1 or PAR2 overexpression in transgenic seedlings are also influenced by prolonged simulated shade treatments but in opposite ways; whereas simulated shade-treated plants are typically longer and paler, those overexpressing PAR1 or PAR2 are shorter and darker than the wild type (Figures 2, 3 and 4). In addition, reduction of PAR1 and/or PAR2 levels results in enhanced elongation phenotypes, suggestive of a mild constitutively active SAS (Figures 5 and 6). Reduced PAR1 and PAR2 transcript levels also result in a severe reduction in seed production (Figure 5C), a trait reported to be similarly affected by simulated shade treatments (Smith and Whitelam, 1997). The negative correlation between PAR1 and PAR2 levels, and the developmental and metabolic responses to simulated shade suggest that PAR1 and PAR2 are negative regulators of SAS.
On the other hand, some of the phenotypes of PAR1 and PAR2 overexpression lines resemble those of auxin, BR or gibberellin mutants (reviewed in Halliday and Fankhauser, 2003). Both gain- and loss-of-function PAR1 and PAR2 plants display altered phenotypes under both W and W+FR (Figures 3, 5 and 6), suggesting that such phenotypes might not be strictly photomorphogenic. Actually, PAR1 and PAR2 may not be considered photomorphogenic components since they are not participating in events from light perception to the first changes in gene expression elicited by the shade signal (i.e., pretranscriptional events). PAR1 and PAR2 are, instead, early components of the shade-regulated transcriptional network, together with ATHB2, HFR1 and PIL1. In this sense, we claim that these genes are components of shade signaling. Simulated shade not only affects the expression of PAR1 and PAR2, but also of a wide diversity of both negative and positive regulators of SAS and, in this networked context, mutants deficient in a single early or primary target of phytochrome action, such as those knocked down in this report, are expected to have a mild effect on the studied phenotypes. When analyzing the role of early components of the transcriptional network initiated by the phytochromes during seedling de-etiolation, similar mild effects were reported (Khanna et al, 2006).
HFR1, a reported master negative regulator of SAS responses, was shown to have a very strong and clear effect under shade conditions that reduced both R:FR ratio and photosynthetic active radiation (amount of light in the 400–700 nm range; Sessa et al, 2005), mimicking natural situations when canopy closure occurs. Our simulated shade conditions, by contrast, only reduce R:FR ratio, without significantly affecting photosynthetic active radiation, mimicking plant proximity detection before actual canopy shading occurs. Under our conditions, the loss-of-function hfr1-5 mutant displays a moderate phenotype, which is qualitatively and quantitatively very similar to that displayed by lines with reduced PAR1 and PAR2 levels (Figures 5 and 6), supporting the fact that all these factors are negative regulators of SAS. The phenotypes resulting from PAR1 and PAR2 overexpression, however, are different from those described in plants overexpressing HFR1, which show clear effects on several light-regulated traits, displaying shorter hypocotyls and higher anthocyanin accumulation than wild-type seedlings after de-etiolation (Yang et al, 2003, 2005; Duek et al, 2004). By contrast, overexpression of PIL1, another negative regulator of SAS, has not been reported to constitutively inhibit elongation responses or pigment accumulation (Salter et al, 2003). The distinct effect of increased levels of HFR1, PIL1, PAR1 or PAR2 on plant development suggests that these factors might control distinct circuits of the shade-regulated transcriptional network involved in implementing SAS responses.
Although no nuclear localization signals in PAR1 or PAR2 could be identified by any web-based program, fusing PAR1 and PAR2 to reporter proteins showed that both are nuclear proteins (Figure 7A). In addition, the characteristic dwarf phenotype of PAR1-overexpressing lines required nuclear targeting of the protein (Figure 7B). These data demonstrate that nuclear localization is required for PAR1 activity, consistent with a role for these atypical bHLHs as transcriptional regulators. As shown here, PAR1 and PAR2 seem to regulate in vivo the transcription of a subset of auxin-regulated genes, including SAUR15 and SAUR68 but not HAT2 (Figure 8). The large proportion of genes misregulated in P35S:PAR1-GG seedlings that are also differentially expressed in wild-type plants upon treatment with auxin and/or BR hormones (Supplementary Figure S5) suggest a broad role for PAR1 (and likely PAR2) in integrating light and hormone signaling networks during SAS. The repressor effect of increased PAR1 levels on auxin-induced SAUR expression is very likely direct, since it is CHX-independent and was observed only 4 h after DEX application (Figure 9B). This time of action is consistent with that reported for other transcription factor-GR fusions over their direct targets (Sablowski and Meyerowitz, 1998; Ohgishi et al, 2001; Roig-Villanova et al, 2006). On the other hand, additional factors might have a more prominent role in the observed decrease in SAUR response to 2,4-D treatment when PAR1 and/or PAR2 levels are constitutively reduced (Figure 8B, data not shown).
Auxin- and BR-signaling pathways converge at the level of transcriptional regulation of target genes with common regulatory elements (reviewed in Halliday, 2004), two of these dual targets being SAUR15 and SAUR68 (Supplementary Table S2). Analysis of only auxin-regulated, and common auxin- and BR-regulated promoters identified G-box and E-box elements, respectively (Nemhauser et al, 2004), both of them recognized by at least some bHLH members. Consistent with SAUR15 being a direct target of PAR1, it has been shown that three E-box sequences present in SAUR15 promoter are necessary for its activation by members of the bHLH family (Yin et al, 2005). Although there is no molecular or biochemical characterization of any member of bHLH group VIII (to which PAR1 and PAR2 belong), they have been suggested to lack the ability to bind DNA, and to form heterodimers, acting as negative regulators of other transcription factors, particularly bHLH proteins (Atchley and Fitch, 1997; Heim et al, 2003; Toledo-Ortiz et al, 2003). It is therefore tempting to speculate that the molecular mechanism behind this repressor effect is the ability of PAR1 and PAR2 to inhibit the DNA-binding activity of transcription activators, most likely belonging to the bHLH family, after heterodimerizing with them. This would make PAR1 and PAR2 transcription cofactors, with the ability to regulate transcription but lacking a DNA-binding domain (Wray et al, 2003).
After simulated shade perception, light signaling networks intersect with those of plant hormones. Auxins are known to play a role in plant responses to changes in light quality (Steindler et al, 1999; Tian and Reed, 2001; Halliday and Fankhauser, 2003). Following auxin transport and accumulation, auxin responsive genes (including Aux/IAA, GH3 and SAUR genes) are induced, triggering signaling pathways (transcriptional cascades) that ultimately lead to cell expansion. Interestingly, simulated shade also affects rapidly but transitorily the expression of SAUR15 and SAUR68. Changes in the expression of SAUR genes in response to auxin can be a marker of auxin-sensitivity to exogenous and, by extension, to endogenous auxin. Our data showing a decreased response to 2,4-D treatment when PAR1 or PAR2 levels are altered can therefore be interpreted as a fine modulation of auxin sensitivity by PAR1 and PAR2. The direct control of PAR gene expression by phytochromes after SAS induction could therefore represent a mechanism to rapidly modulate some auxin responses (SAUR expression) and to integrate shade perception and hormone signaling pathways.
Materials and methods
Plant material and growth conditions
Arabidopsis (A. thaliana) transgenic lines were generated in the Col-0 background. The SALK_109270 line was named par2-1. The mutant lines hfr1-4 and hfr1-5, also generated by the SALK collection (Alonso et al, 2003), have been described previously (Sessa et al, 2005). Details of the genotyping of hfr1 and par2-1 mutant plants are given in the Supplementary data. Adult plants were either grown under short-day photoperiodic conditions (Figure 2A), or in the greenhouse (Figure 5C), as described (Martínez-García et al, 2002). All the other experiments were performed with seedlings grown in plates, as detailed below. Seeds were germinated on Petri dishes with solid growth medium (without sucrose, GM−) as described (Roig-Villanova et al, 2006). Plates were incubated in a I-36VL growth chamber (Percival Scientific Inc., Perry, IA, USA) at 22°C under two different light conditions: (i) W, which was provided by four cool-white vertical fluorescent tubes (80 μmol m−2 s−1 of photosynthetically active radiation; R:FR ratio of 3.2–4.5) and (ii) simulated shade (W+FR), which was generated by enriching W with supplementary FR provided by QB1310CS-670-735 LED hybrid lamps (Quantum Devices Inc., Barneveld, WI, USA) (80 μmol m−2 s−1 of photosynthetically active radiation; R:FR ratio of 0.05). Fluence rates were measured using an EPP2000 spectrometer (StellarNet Inc., Tampa, FL, USA).
Construction of transgenic lines
Details of the generation of the constructs used to obtain the described transgenic lines are provided in the Supplementary data. Arabidopsis plants were transformed with the obtained binary vectors as described (Roig-Villanova et al, 2006). The presence of the transgene in the selected T1 plants was verified by PCR analysis. Only lines with a single T-DNA insertion (as estimated from the segregation of the marker gene in T2 populations) were eventually selected.
RNA blot analysis
Total RNA was isolated from seedlings, electrophoresed and blotted as described (Roig-Villanova et al, 2006). Hybridization probes for PAR1, HAT2 and 25S rRNA were prepared as described (Roig-Villanova et al, 2006). Probes for PAR2, SAUR15 and SAUR68 were made from the respective full-length fragments cloned in pJB3, pAG3 and pAG2, by PCR using specific primers (see Supplementary data). Expression levels were normalized with the 25S rRNA signal. Hybridization, washes, exposure and quantification of radioactive signals were carried out as described (Martínez-García et al, 2002). These experiments were conducted at least twice.
Physiological measurements
The National Institutes of Health ImageJ software (Bethesda, MD, USA) was used on digital images to quantify hypocotyl length (after laying out seedlings flat on agar plates) and cotyledon and primary leaf longitudinal expansion (after rolling seedlings flat on a transparent self-adhesive sheet). At least 15 seedlings were used for each treatment and experiments were repeated 2–5 times. Data shown in Figures 3, 5B, 6C and D, correspond to a representative experiment. Error bars represent twice the standard error of the mean (2 × s.e.), which corresponds to 95% confidence intervals (Cumming et al, 2007). Photosynthetic pigments were extracted, separated by HPLC and quantified as indicated (Rodríguez-Concepción et al, 2004). These experiments were repeated twice and a representative experiment is shown (Figure 4). Statistical analysis of the data was performed using the Simple Interactive Statistical Analysis (SISA) T-test available online (http://home.clara.net/sisa/t-test.htm). Two-way ANOVA tests were performed using GraphPad Prism version 4.00 for Windows (GraphPad Software, San Diego, CA, USA).
CHX, DEX and 2,4-D treatments
CHX (Sigma-Aldrich) was dissolved in 50% (v/v) ethanol at 50 mM; DEX and 2,4-D (Sigma-Aldrich) were dissolved in 100% ethanol (v/v) at 5 and 250 mM, respectively. These stock solutions were kept at −20°C until use. Working solutions were prepared in water prior to the treatments. Treatments were performed using 7-day-old seedlings grown on filter-paper circles, as described (Roig-Villanova et al, 2006).
Sequence and phylogenetic analysis
Multiple sequence alignments were performed using the CLUSTALX 1.8 program (Thompson et al, 1997). Alignments were edited with GeneDoc (www.psc.edu/biomed/genedoc). Limits of the bHLH domains were taken according to the proposed consensus motif (Ferre-D'Amare et al, 1993; Atchley et al, 1999; Toledo-Ortiz et al, 2003). Neighbor joining and 50% majority-rule consensus trees were constructed using NEIGHBOR and CONSENSUS, respectively from the PHYLIP package (evolution.gs.washington.edu/phylip.html). To provide statistical confidence on the retrieved topology, a bootstrap analysis of 100 replicates was performed through the SEQBOOT application. The trees were represented using the TreeView v1.6.6. software (Page, 1996).
Subcellular localization analysis
Transgenic seedlings were mounted in water on glass slides. GFP fluorescence was inspected with a Leica TCS SP confocal microscope using a 488 nm argon laser-line (Leica Microsystems, Heidelberg, Germany). At least two independent transgenic lines were examined for each construct.
Supplementary Material
Supplementary Data
Acknowledgments
We thank the Servei d'Hivernacles for plant care; A Carbonell, B Freijomil, L Rodríguez and M Galiáñez for help and support; Gloria Garcia and Roberto Solano (Genomic Facility, GEFA Project, CNB, Madrid) for performing the transcriptomic analysis and advise on data presentation; and J Casacuberta, P Devlin and P Más for comments on the manuscript. IR-V and AG received an FPI and FPU fellowship from the Spanish Ministry of Science and Education (MEC), respectively. JB-T financial support came from the Generalitat de Catalunya (GC) (2004CRED-10003, 2005CRED-00014). Our research is supported by grants from the GC and Spanish MEC-FEDER to JFM-G (BIO2002-00298; BIO2005-00154, 2005SGR-00284 and Xarxa de Referència en Biotecnologia) and MRC (BIO2005-00367, 2005SGR-00914 and Distinció de la Generalitat).
Accession number Microarray data were deposited with MIAMExpress under accession number E-ATMX-29.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM (1997) A natural classification of the basic helix–loop–helix class of transcription factors. Proc Natl Acad Sci USA 94: 5172–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Terhalle W, Dress A (1999) Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J Mol Evol 48: 501–516 [DOI] [PubMed] [Google Scholar]
- Blanc G, Barakat A, Guyot R, Cooke R, Delseny M (2000) Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MJ, Atchley WR (2003) Phylogenetic analysis of plant basic helix–loop-–helix proteins. J Mol Evol 56: 742–750 [DOI] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I (1996) Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA 93: 3530–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C (2004) Light signal transduction in higher plants. Annu Rev Genet 38: 87–117 [DOI] [PubMed] [Google Scholar]
- Cumming G, Fidler F, Vaux DL (2007) Error bars in experimental biology. J Cell Biol 177: 7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PRH, Patel SR, Goosey L, Sharrock RA, Whitelam GC (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation and flowering time. Plant Physiol 119: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C (2004) The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol 14: 2296–2301 [DOI] [PubMed] [Google Scholar]
- Faigón-Soverna A, Harmon FG, Storani L, Karayekov E, Staneloni RJ, Gassmann W, Más P, Casal JJ, Kay SA, Yanovsky MJ (2006) A constitutive shade-avoidance mutant implicates TIR-NBS-LRR proteins in Arabidopsis photomorphogenic development. Plant Cell 18: 2919–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Prendergast GC, Ziff EB, Burley SK (1993) Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363: 38–45 [DOI] [PubMed] [Google Scholar]
- Halliday KJ (2004) Plant hormones: the interplay of brassinosteroids and auxin. Curr Biol 14: R1008–R1010 [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Fankhauser C (2003) Phytochrome-hormonal signaling networks. New Phytol 157: 449–463 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hensel LL, Nelson MA, Richmond TA, Bleecker AB (1994) The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol 106: 863–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH (2004) PHYTOCHROME-INTERACTING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH (2002) PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J 21: 2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH (2004) A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic–helix–loop–helix transcription factors. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Toledo-Ortiz G, Kikis EA, Johannesson H, Hwang YS, Quail PH (2006) Functional profiling reveals that only a small number of phytochrome-regulated early-response genes in Arabidopsis are necessary for optimal deetiolation. Plant Cell 16: 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G (2003) Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH (2000) Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Virgós-Soler A, Prat S (2002) Control of photoperiod-regulated tuberization in potato by the Arabidopsis flowering-time gene CONSTANS. Proc Natl Acad Sci USA 99: 15211–15216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T (1987) Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol 9: 611–623 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ohgishi M, Oka A, Morelli G, Ruberti I, Aoyama T (2001) Negative autoregulation of the Arabidopsis homeobox gene ATHB-2. Plant J 25: 389–398 [DOI] [PubMed] [Google Scholar]
- Page RD (1996) TreeView: an application to display phylogenetic trees on personal computers. Comp App Biosciences 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martínez-García JF (2006) Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol 141: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Concepción M, Forés O, Martínez-García JF, Gozález V, Phillips MA, Ferrer A, Boronat A (2004) Distinct light-mediated pathways regulate the biosynthesis and exchange of isoprenoid precursors during Arabidopsis seedling development. Plant Cell 16: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski RWM, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an intermediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103 [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sawa S, Ohgishi M, Goda H, Higuchi K, Shimada Y, Yoshida S, Koshiba T (2002) The HAT2 gene, a member of the HD-ZIP gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J 32: 1011–1022 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H (1982) Light quality, photoperception, and plant strategy. Annu Rev Plant Physiol 33: 481–518 [Google Scholar]
- Smith H, Whitelam GC (1997) The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ 20: 840–844 [Google Scholar]
- Steindler C, Matteuci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW (2001) Molecular links between light and auxin signaling pathways. J Plant Growth Regul 20: 274–280 [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix–loop–helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20: 1377–1419 [DOI] [PubMed] [Google Scholar]
- Yang J, Lin R, Sullivan J, Hoecker U, Liu B, Xu L, Deng XW, Wang H (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17: 804–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KY, Kim YM, Lee S, Song PS, Soh MS (2003) Overexpression of a mutant basic helix–loop–helix protein HFR1, HFR1-deltaN105, activates a branch pathway of light signaling in Arabidopsis. Plant Physiol 133: 1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis Microarray Database and Analysis Toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data