Abstract
Although the mechanisms for regulation of ribosomal protein gene expression have been established for gram-negative bacteria such as Escherichia coli, the regulation of these genes in gram-positive bacteria such as Bacillus subtilis has not yet been characterized. In this study, the B. subtilis rpsD gene, encoding ribosomal protein S4, was found to be subject to autogenous control. In E. coli, rpsD is located in the alpha operon, and S4 acts as the translational regulator for alpha operon expression, binding to a target site in the alpha operon mRNA. The target site for repression of B. subtilis rpsD by protein S4 was localized by deletion and oligonucleotide-directed mutagenesis to the leader region of the monocistronic rpsD gene. The B. subtilis rpsD leader exhibits little sequence homology to the E. coli alpha operon leader but may be able to form a pseudoknotlike structure similar to that found in E. coli.
Full text
PDF

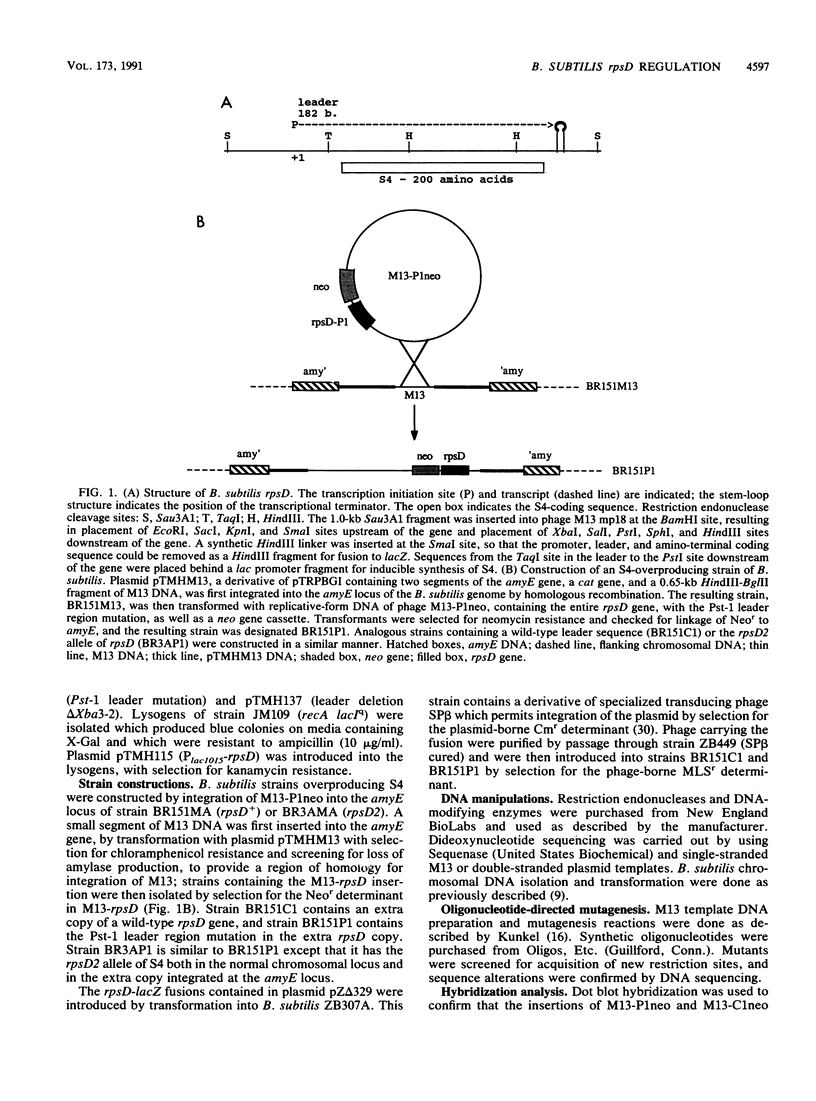

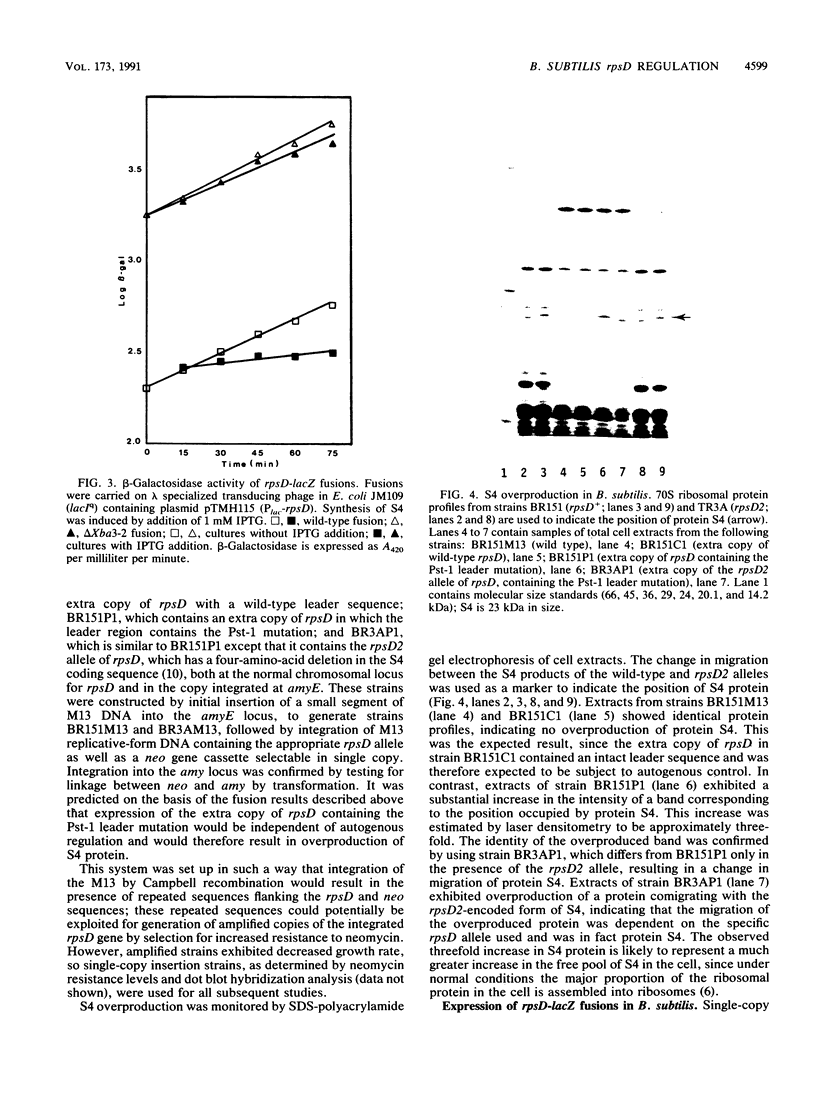

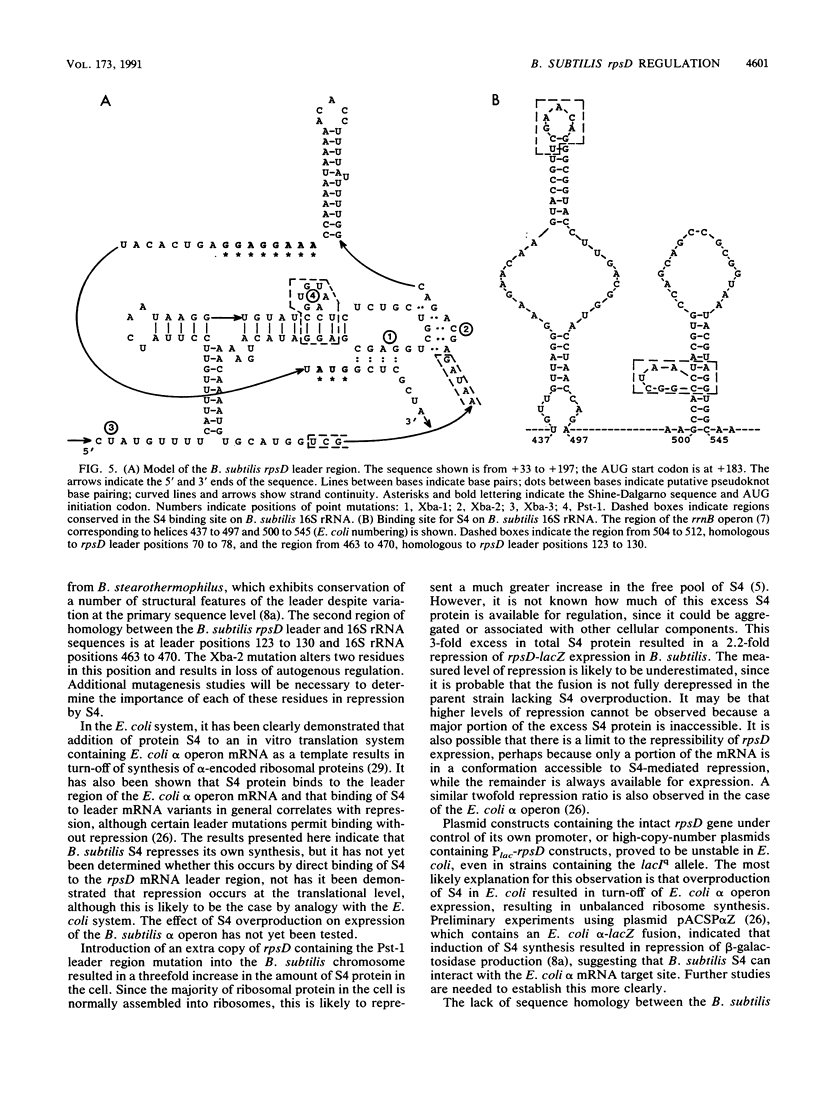

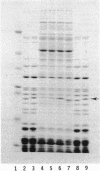
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bedwell D., Davis G., Gosink M., Post L., Nomura M., Kestler H., Zengel J. M., Lindahl L. Nucleotide sequence of the alpha ribosomal protein operon of Escherichia coli. Nucleic Acids Res. 1985 Jun 11;13(11):3891–3903. doi: 10.1093/nar/13.11.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohannon D. E., Sonenshein A. L. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol. 1989 Sep;171(9):4718–4727. doi: 10.1128/jb.171.9.4718-4727.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Suh J. W., Thomas S. M., Price C. W. Gene encoding the alpha core subunit of Bacillus subtilis RNA polymerase is cotranscribed with the genes for initiation factor 1 and ribosomal proteins B, S13, S11, and L17. J Bacteriol. 1989 May;171(5):2553–2562. doi: 10.1128/jb.171.5.2553-2562.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D., Nomura M. Feedback regulation of ribosomal protein gene expression in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3590–3594. doi: 10.1073/pnas.77.6.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gausing K. Ribosomal protein in E. coli: rate of synthesis and pool size at different growth rates. Mol Gen Genet. 1974 Mar 6;129(1):61–75. doi: 10.1007/BF00269266. [DOI] [PubMed] [Google Scholar]
- Green C. J., Stewart G. C., Hollis M. A., Vold B. S., Bott K. F. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon, rrnB. Gene. 1985;37(1-3):261–266. doi: 10.1016/0378-1119(85)90281-1. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. Cloning and analysis of the Bacillus subtilis rpsD gene, encoding ribosomal protein S4. J Bacteriol. 1990 Nov;172(11):6372–6379. doi: 10.1128/jb.172.11.6372-6379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin T. M., Chambliss G. H. Genetic mapping of a mutation causing an alteration in Bacillus subtilis ribosomal protein S4. Mol Gen Genet. 1984;193(2):364–369. doi: 10.1007/BF00330694. [DOI] [PubMed] [Google Scholar]
- Henkin T. M., Chambliss G. H., Grundy F. J. Bacillus subtilis mutants with alterations in ribosomal protein S4. J Bacteriol. 1990 Nov;172(11):6380–6385. doi: 10.1128/jb.172.11.6380-6385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin T. M., Sonenshein A. L. Mutations of the Escherichia coli lacUV5 promoter resulting in increased expression in Bacillus subtilis. Mol Gen Genet. 1987 Oct;209(3):467–474. doi: 10.1007/BF00331151. [DOI] [PubMed] [Google Scholar]
- Hori H., Higo K., Osawa S. The rates of evolution in some ribosomal components. J Mol Evol. 1977 May 13;9(3):191–201. doi: 10.1007/BF01796108. [DOI] [PubMed] [Google Scholar]
- Itaya M., Kondo K., Tanaka T. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 1989 Jun 12;17(11):4410–4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S., Nomura M. Ribosomal protein S4 acts in trans as a translational repressor to regulate expression of the alpha operon in Escherichia coli. J Bacteriol. 1982 Jul;151(1):193–202. doi: 10.1128/jb.151.1.193-202.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nakano M. M., Zuber P. Cloning and characterization of srfB, a regulatory gene involved in surfactin production and competence in Bacillus subtilis. J Bacteriol. 1989 Oct;171(10):5347–5353. doi: 10.1128/jb.171.10.5347-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Smith D. K., Olsen G. J., James B. D. Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA--a review. Gene. 1989 Oct 15;82(1):65–75. doi: 10.1016/0378-1119(89)90031-0. [DOI] [PubMed] [Google Scholar]
- Roland K. L., Liu C. G., Turnbough C. L., Jr Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7149–7153. doi: 10.1073/pnas.85.19.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43(1-2):85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Stern S., Wilson R. C., Noller H. F. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986 Nov 5;192(1):101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- Stoker N. G., Fairweather N. F., Spratt B. G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982 Jun;18(3):335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Evidence for allosteric coupling between the ribosome and repressor binding sites of a translationally regulated mRNA. Biochemistry. 1990 May 8;29(18):4434–4439. doi: 10.1021/bi00470a025. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Vartikar J. V., Draper D. E. S4-16 S ribosomal RNA complex. Binding constant measurements and specific recognition of a 460-nucleotide region. J Mol Biol. 1989 Sep 20;209(2):221–234. doi: 10.1016/0022-2836(89)90274-x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Arfsten A. E., Nomura M. In vitro expression of Escherichia coli ribosomal protein genes: autogenous inhibition of translation. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1837–1841. doi: 10.1073/pnas.77.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber P., Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987 May;169(5):2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]