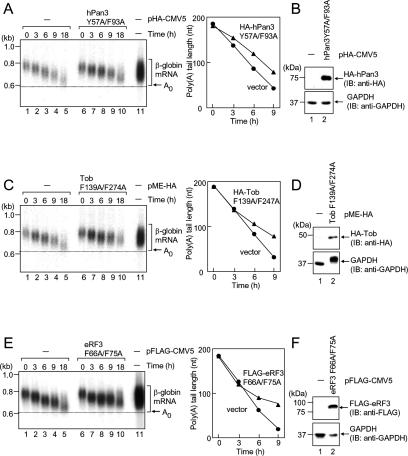
Figure 7.
hPan3, Tob, and eRF3 with mutations in the PABPC1-binding PAM2 motifs affect mRNA deadenylation in a dominant-negative fashion. (A) T-Rex-HeLa cells were transfected with pFlag-CMV5/TO-G1 reporter and either pHA-CMV5 (lanes 1–5) or pHA-hPan3 Y57A/F93A (lanes 6–10). Transcriptional pulse-chase analysis was performed as described in Figure 2. (B) Total cell lysates were analyzed by immunoblotting using anti-HA (top) and anti-GAPDH (bottom). (C) As in A, except that the cells were transfected with pMEHA or pMEHA–Tob F139A/F274A, and transcriptional pulse-chase analysis was performed. (D) As in B, total cell lysates were immunoblotted. (E) As in A, except that the cells were transfected with pFlag-CMV5 or pFlag-eRF3 F66A/F75A, and transcriptional pulse-chase analysis was performed. (F) As in B, except that anti-Flag antibody was used, and total cell lysates were immunoblotted.