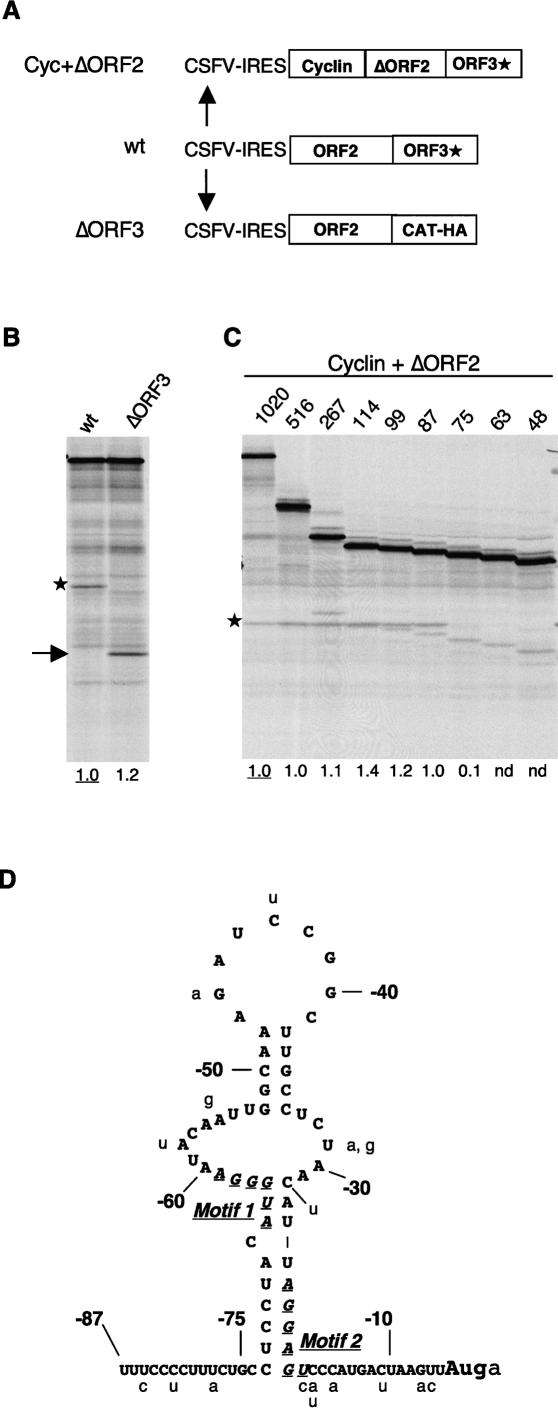
Figure 3.
Mapping the viral sequences required for downstream ORF expression. (A) Schematic diagram (not to scale) of the deletions introduced to map the requirement for viral sequences. (B) Translation of the ΔORF3 mRNA shows that ORF3 coding sequences are not required for downstream cistron expression. The full-length ORF3* product is indicated by an asterisk, and the CAT-HA product is indicated by an arrow. (C) Deletion mapping of the ORF2 sequences required for ORF3* expression. The deletions are designated according to the number of 3′-terminal residues of ORF2 retained. The ORF3* product is indicated by an asterisk. Numbers below each lane show the efficiency of reinitiation (the ORF3*/ORF2 product yield ratio) expressed relative to the control mRNA value set at 1.0. (nd) Not detectable above background (i.e., effectively zero). The anomalous high value for the construct with 114 nt retained is due to the comigration of the ORF3* product with an incomplete translation product of ORF2. (D) Sequence and tentative secondary structure prediction of the 3′-terminal 87 nt of FCV strain F9 ORF2, with the variations found in other strains shown in lowercase. The core segments of Motif 1 and Motif 2 identified by Luttermann and Meyers (2007) as especially critical for downstream cistron translation are highlighted by italics and underlining.