Figure 1.
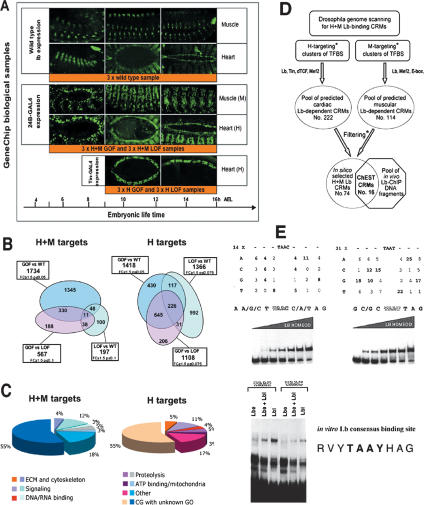
Identification of Lb target genes by expression profiling and ChEST strategies. (A). Muscle- and heart-specific expression of lb and the two drivers (24B > GFP and tin > GFP) used for targeted transcriptional profiling. Three different embryonic stages are presented to illustrate Lb and 24B-GAL4 expressions in biological samples used for microarray experiments. In the first column (5–9 h AEL) are shown embryos at stage 11 or early stage 12 (lateral views). In the middle column (9–12 h AEL) are embryos at stage 13, and in the third column (12–16 h AEL) are shown embryos at stage 15–16. In the middle and right column, muscle (M) expression is illustrated by lateral views and heart (H) expression by dorsal views of embryos. Note that tin-GAL4 driver expression starts later, from stage 13, and persists until the late embryogenesis. To obtain wild-type biological samples for microarray and ChEST experiments, three collections of embryos aged from 5 to 16 h AEL have been used. In a similar manner, F1 embryos from the cross of 24B-GAL4 with UAS-lbe and with UAS-lbRNAi lines were collected and used as H + M GOF and H + M LOF samples, respectively. Embryos from the cross of tin-GAL4 with UAS-lbe or UAS-lbRNAi lines were aged according to tin-GAL4 expression and later labeled H GOF and H LOF, respectively. (B) Venn diagrams showing populations of candidate genes identified by comparison of GOF versus wild-type, LOF versus wild-type, and GOF versus LOF context. Note that in H conditions more candidate genes are common for these three candidate gene populations than in H + M context. (C) Main categories of Lb targets identified by targeted expression profiling. GO-based statistical distribution of genes identified in H + M targeting conditions and H targeting conditions. In both contexts we find an important enrichment for ECM and cytoskeleton components, signaling molecules, proteins involved in proteolysis, ATP binding, and factors carrying a DNA- or RNA-binding domain. Note that >50% of identified genes have no GO annotations and ∼17% have a GO, which does not fit into any of the five main categories. (D) Genome scanning strategy for Lb-binding CRMs. Different heart (H) targeting and muscle (M) targeting genome scanning conditions were used (asterisk; see Supplemental Material). A pool of predicted cardiac and muscular Lb-dependent CRMs was filtered with respect of distance to adjacent genes and their annotations (asterisk; see Supplemental Material). Selected CRMs were spotted to produce a computed Lb-CRM array. In parallel, a pool of DNA fragments bound in vivo by Lb was isolated by ChIP and used to probe the computed array. Among 74 CRMs spotted, 16 were found enriched in ChIP material. (E) Identification of the in vitro consensus binding site for Ladybird. Random 10-nt-long oligos flanked on 5′ and 3′ by 20-nt primer-compatible sequences were radiolabeled and used for the SELEX approach. Four cycles of incubation with 6xHis-Lb homeodomain fusion protein followed by the amplification of a selected subset of sequences were applied to select motifs bound preferentially by Lb. As shown by shift assays, Lb homeodomain (top panels) and both Lbe and Lbl proteins (bottom panel) recognize sequences containing TAAT and TAAC core motifs, and the deduced consensus sequence is RVYTAAYHAG.