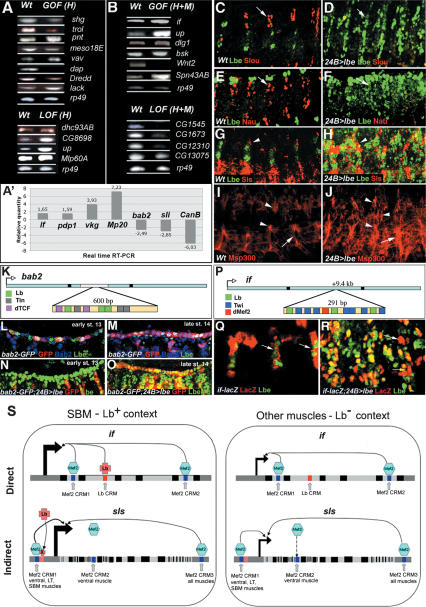
Figure 2.
Validation of selected Lb targets identified by expression profiling and ChEST approaches. (A,B) RT–PCR analyses of transcript levels in wild type and in lb GOF or LOF conditions for candidates identified in H targeting (A) and H + M targeting (B) conditions. At least two candidate genes from each of main categories have been analyzed (see also Table 1A,B). (A′) Real-time RT–PCR validation of additional candidates. Relative quantity of gene transcripts in GOF H + M, LOF H + M, or GOF H contexts versus wild type are presented (see Materials and Methods and Supplementary Table S8 for details). Alterations in candidate gene expression observed in RT–PCR and real-time RT–PCR experiments are consistent with expression changes identified by microarrays. Lateral views of stage 14 (C,D,G–J) and stage 13 (E,F) embryos. lb is able to repress (C–F) certain target genes and to activate (G–J) others. (C–F) Expression of muscle identity gene slou (C) and nau (E) are repressed by lb in 24B > lbe embryos (arrows in D,F). (G–J) Expression of Sls, a component of sarcomeric cytoskeleton (G) and a dystrophin related Msp300 (I) are ectopically activated (arrows in H,J) in lbe GOF conditions. Arrowheads and arrow indicate increased Sls (H) and Msp300 (J) level in the SBM and the ventral muscles, respectively. bab2 (K–O) and if (P–R) CRMs act as Lb-dependent regulatory regions in vivo. (K,P) A scheme showing the position and organization of Lb-dependent CRMs located within the first bab2 (K) and if (P) intron. Positions of Lb, Tin, Twi, Dmef2, and dTCF-binding sites are indicated. (L) Wild-type expression of bab2-GFP line (carrying Lb-dependent CRM) showing that GFP expression is initiated in Lbe cardiac cells at stage 13. Notice that initially in each hemisegment only one out of two Lbe-positive cardioblasts express GFP. GFP expression also coincides with endogenous Bab2 expression. (M) Wild-type bab2-GFP expression in stage 14 embryo showing that during later steps of cardiac development the CRM drives expression in an enlarged population of cardiac cells including Lbe-negative cardioblasts. GFP is coexpressed with endogenous Bab2. (N) bab2-GFP expression is not seen at stage 13 in embryos overexpressing Lbe in mesodermal cells, suggesting that during early stages of heart development lb is able to repress bab2. (O) In stage 14 embryo overexpressing Lbe, bab2-GFP is no longer repressed, indicating that the repressive influence of Lbe on this CRM is transient. (Q) The ChEST-identified if CRM drives LacZ expression in the SBM (arrows). (R) In embryos expressing Lbe ectopically in all muscle cells, expression of LacZ is enlarged (yellow arrow). (S) A scheme showing a model based on identity gene-dependent fine-tuning of muscle gene expression. Both direct (if case) and indirect (sls case) regulatory mechanisms are depicted. Black boxes represent exons, gray boxes represent the upstream and downstream noncoding sequences, and the light-gray boxes correspond to introns. Positions of dMef2 (Junion et al. 2005) and Lb-dependent CRMs are indicated. In pink is labeled a CRM to which binds hypothetical Lb-regulated factor X. if and sls transcription is regulated by generic dMef2-dependent modules. In the SBM context (in the presence of Lb protein) Lb binds to its intronic if CRM and contributes to the regulation of if transcription. In the case of sls, Lb action is indirect via an as-yet-unknown factor X. This factor most probably contributes to the activity of dMef2-dependent CRM1, which drives expression in a subset of muscles including SBM (Junion et al. 2005).