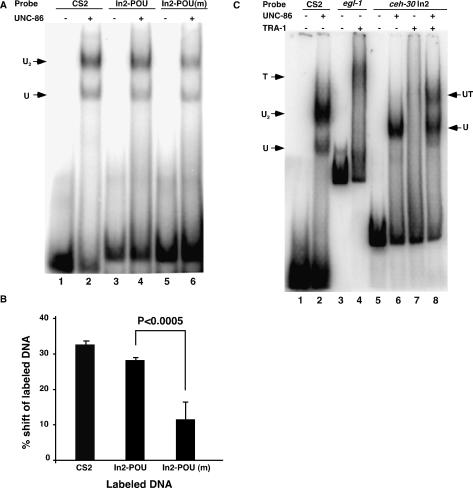
Figure 3.
Both UNC-86 and TRA-1A bind to the intron 2 sequence of ceh-30 in vitro. (A) Binding of GST∷UNC-86 to a POU-type homeodomain-binding site (In2-POU) found in intron 2 of ceh-30 and an altered site [In2-POU(m)] derived from the sm130 mutant. Twenty-five nanograms of recombinant GST∷UNC-86 (lanes 2,4,6) or binding buffer alone (lanes 1,3,5) were incubated with labeled oligonucleotides derived from a known UNC-86-binding site (CS2; lanes 1,2), In2-POU (lanes 3,4), or In2-POU(m) (lanes 5,6) and resolved on an 8% native polyacrylamide gel. (U) The gel shift species containing the UNC-86 monomer; (U2) the gel shift species containing the UNC-86 homodimer (Xue et al. 1993). (B) The sm130 lesion reduces the binding of GST∷UNC-86 to In2-POU. The fraction of labeled probe shifted by GST∷UNC-86 was quantified from four independent experiments as described in Materials and Methods. (C) UNC-86 stabilizes the binding of TRA-1A to the intron 2 sequence of ceh-30. Twenty-five nanograms of recombinant GST∷UNC-86 and 5 ng of TRA-1∷His6 either alone or together were incubated with labeled CS2 (lanes 1,2), 257-bp DNA fragment containing the TRA-1A-binding site from the egl-1 gene (egl-1, lanes 3,4), or 91-bp intron 2 DNA fragment from ceh-30 containing both In2-POU and In2-TRA (ceh-30 In2, lanes 5–8). The reactions were resolved on a 4% native polyacrylamide gel. (T) The gel shift species containing the TRA-1 protein; (UT) the gel shift species containing both UNC-86 and TRA-1A.