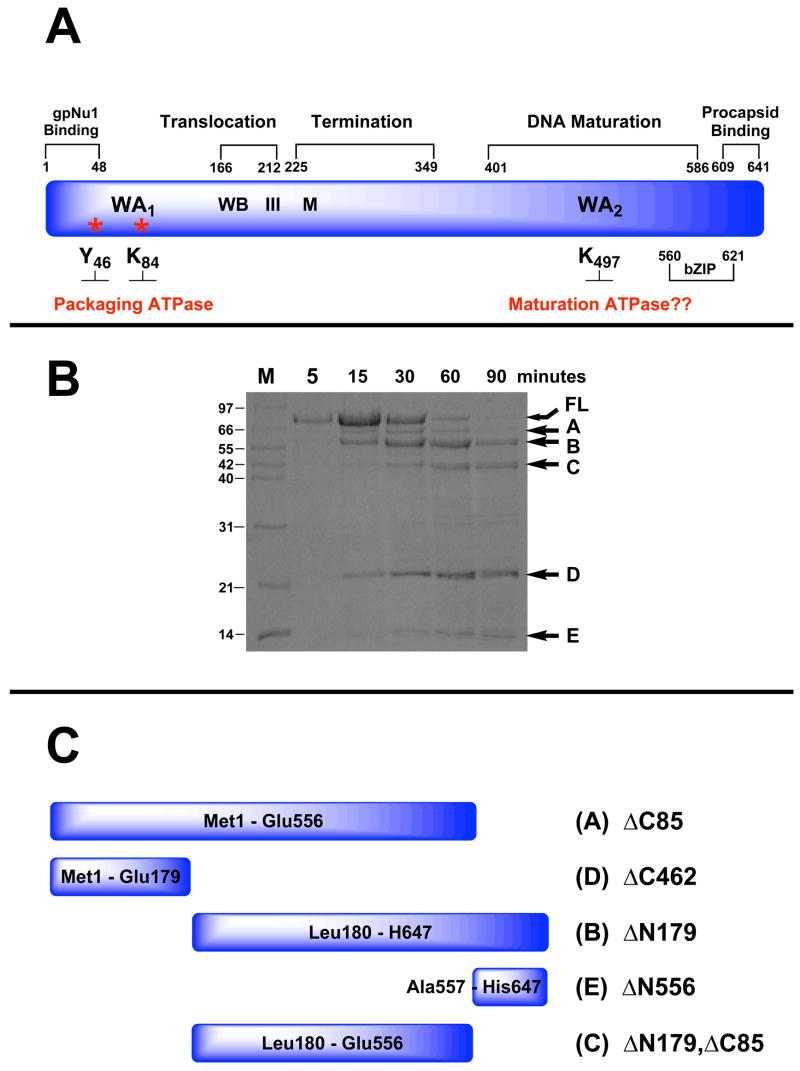
Figure 2.
Panel A. Genetic Organization of gpA. The full-length protein (641 residues) is depicted in blue and functional domains identified by genetic studies are indicated above. Walker A and B sequences (WA1, WB) identified by primary sequence analysis and associated with the packaging ATPase site are indicated. Residues covalently modified with AzATP (Y46, K84) are indicated with asterisks. Helicase motif III and metal binding (M) residues identified by primary sequence analysis are also indicated. The putative Walker A sequence in the C-terminal DNA maturation ATPase site (WA2 identified by primary sequence analysis) is also indicated. Panel B. Limited Proteolysis of gpA by Endo-Glu-C. The isolated gpA subunit was digested with Endo-Glu-C as described in Materials and Methods and the time course monitored by SDS-PAGE. Lane M, protein molecular weight standards with sizes indicated to the left of the gel. The undigested, full-length protein (FL) and protease resistant fragments (A–E) are indicated with arrows at right. Panel C. Cartoon depicting the deduced sequence of the gpA protease-resistant peptides.