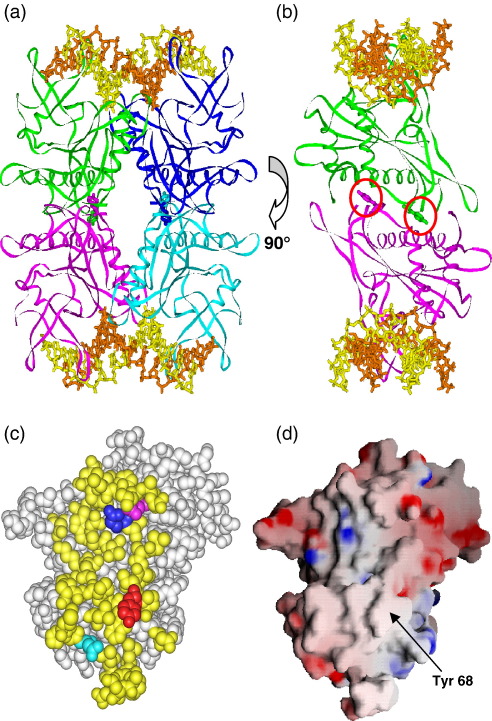
Fig. 1.
SfiI structures. (a) The crystal structure of tetrameric SfiI endonuclease bound to two specific DNA duplexes (RCSB Protein Data Bank, accession code 2EZV).25 Two polypeptide chains (represented as green and blue ribbons) form a primary dimer bound to one DNA, and the other two (magenta and cyan ribbons) form a second dimer bound to another DNA. In both duplexes, the two strands are in orange and yellow. (b) Side-on view. The structure of the SfiI–DNA complex in (a) has been rotated through 90°, and the two monomers in blue and cyan, which would be at the front of this view, have been removed for clarity. In both (a) and (b), the Tyr68 residues at the interdimer interface are shown in ball-and-stick format and, in (b), are highlighted with red circles. (c) Space-filling representation of the surface of the magenta-coloured monomer in (b) that faces the monomer in green across the dimer interface. All of the residues in yellow lie within 5 Å of the green monomer. Residues that make side-chain-to-side-chain interactions with the opposite monomer are coloured as follows: Tyr68, red; Gln30, purple; Gln3, light blue; Gln26, dark blue. (d) Electrostatic potential48 on the protein surface shown in (c). Positively charged residues are in blue, negatively charged residues are in red and uncharged residues are in white. The arrow marks Tyr68.