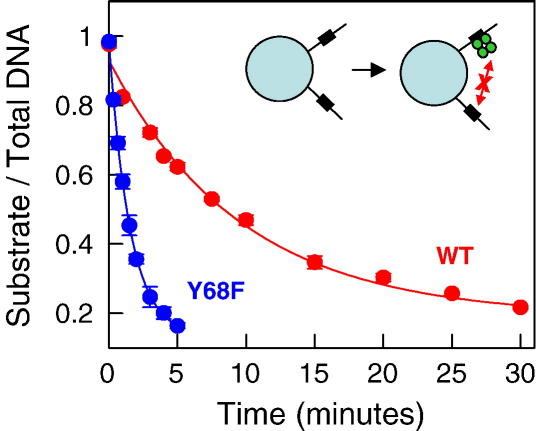
Fig. 7.
Immobilised oligonucleotides. Samples at 50 °C contained, in reaction buffer without MgCl2, SfiI protein (50 nM) and 32P-labelled BIO-30 (final concentration, 1 nM) that had been previously bound to streptavidin-coated magnetic beads (125 nM in streptavidin). The cartoon shows two specific duplexes (rectangles to mark recognition sites) bound to a bead (cyan circle) that are too far apart to allow the SfiI tetramer (green circles) to bridge the duplexes. The protein was either wt SfiI (red circles) or the Y68F mutant (blue circles). MgCl2 was then added to a final concentration of 10 mM. Aliquots were taken at the times indicated on the x-axis and analysed as in Materials and Methods to determine the extent of cleavage of the labelled strand of BIO-30. The amount of intact DNA, relative to the total, is given on the y-axis. The line through each set of data points is the best fit to a first-order rate constant: for wt SfiI, 0.1 min− 1; for Y68F, 0.7 min− 1.