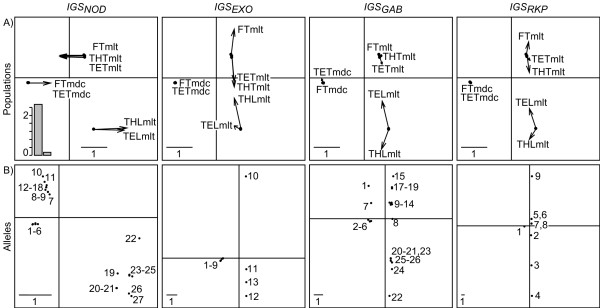
Figure 6.
Application of the DPCoA-MCoA to the real data set. A) Comparison between the patterns of the differences among populations given by the compromise over all loci (black dots, start of the arrows) and the individual analyses (end of the arrows). The special status of IGSNOD is highlighted by horizontal arrows (wrong assignment on the first axis), whereas IGSGAB, IGSRKP and IGSEXO presents vertical arrows (discrepancies from the compromise structure on axis 2 only); B) Location of the alleles. A low (or high) variance in allele points on an axis indicates that the diversity among alleles within populations is lower (or higher) than the diversity among populations, because each axis is normalized for diversity among populations. An eigenvalue barplot is provided in the left-hand corner.