Figure 1.
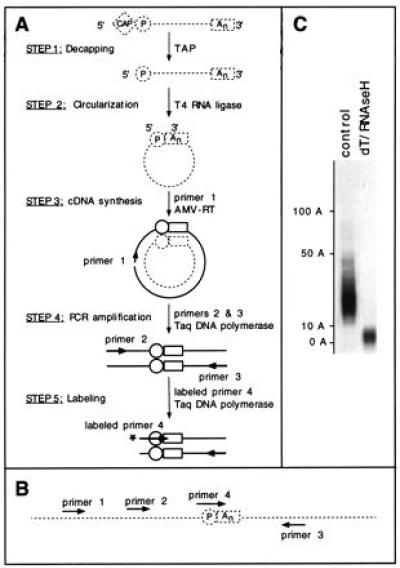
The cRT-PCR procedure. (A) Strategy used for measuring the poly(A) tail length of a specific mRNA. Messenger RNA molecules are represented as broken lines and DNA molecules as thick lines. The cap and poly(A) tail (An) are indicated by a diamond and a rectangle, respectively. The circle indicates the 5′-phosphate group of the most 5′ base of the original mRNA. AMV, avian myeloblastosis virus. (B) Arrangement of the primers used. The junction point of the circularized RNA is shown. (C) cRT-PCR allows one to measure the size distribution of the poly(A) tail. Albumin (ALB) mRNA poly(A) tail length was analyzed using hepatoma cells RNAs. The RNAs were decapped with tobacco acid pyrophosphatase (TAP) and subjected to cRT-PCR without (control lane), or with (dT/RNase H lane) removal of most of the poly(A) tail by treatment with oligo(dT) and RNase H. The length of the poly(A) tails as deduced by comparison to a size standard is indicated to the left. Labeling was performed during the PCR (25 cycles) using labeled primer 3.
