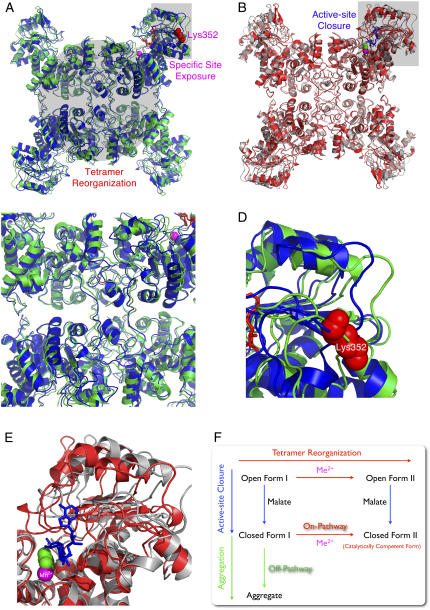
FIGURE 10.
Ligand-induced structural changes of ME. Interconversion of the open form I (pdb code: 1QR6), open form II (pdb code: 1PJL), closed form II (pdb code: 1DO8), and the putative closed form I. (A) Superimposition of the open form I (in blue) and open form II (in green) showing the tetramer reorganization and the specific trypsin-cutting site exposure upon binding of metal ion (in purple sphere). (B) Superimposition of the open form II (in gray) and closed form II (in red) showing the active site closure after substrate binding (here in the figure is the transition state analog oxalate in the green sphere). The superimposition was generated by Swiss-PDBViewer v. 3.7 (38). (C) Zoom-in area showing the tetramer reorganization. (D) Zoom-in area showing the exposure of the specific cutting site in the open form II. (E) Zoom-in area showing the closure of active site. (F) Proposed model for the structural interconversion of ME. The enzyme binds the nucleotide first forming open form I. Mn2+ and L-malate can then be bound in any order forming open form II by tetramer reorganization or closed form I by the active-site closure. Without Mn2+, close form I (the putative molten globule state) is inherently unstable and is prone to divergence into aggregation (off-pathway). Mn2+ plays a protective role by helping the enzyme enter the correct pathway (on-pathway) leading to its catalytically competent closed form II.