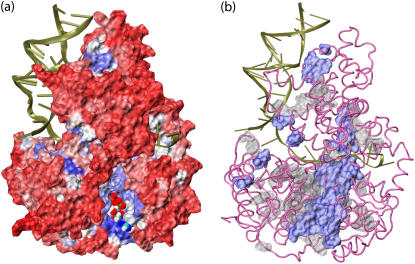
FIGURE 6.
Conserved residues in the PcrA family. Conservation of residues was determined through sequence alignment of a family of proteins; for smaller families more residues are conserved than for larger families. We based our analysis on two families, a large family including 500 sequences of PcrA, Rep, and UvrD helicases (alignment 1), and a small family including 106 sequences of PcrA helicases (from different species; alignment 2). The proteins in the two families were selected through PSI-BLAST (30,31); the alignment was obtained through ClustalW (32). Shown in a is the degree of conservation for PcrA-Rep-UvrD helicases from alignment 1. The PcrA-DNA complex (from BACST) is colored according to the calculated score of conservation using the SCA software package (29): red represents nonconserved residues while blue represents conserved ones; DNA is shown in cartoon presentation, the protein in surface presentation, and the bound-ATP in vdW presentation. One can recognize that the protein region around ATP and DNA exhibits preferentially conserved residues (blue). Shown in b is a comparison of conserved residues for the families of PcrA-Rep-UvrD helicases (blue) and PcrA-only helicases (gray) from alignment 2. It can be noticed that there are many conserved residues near dsDNA in PcrA-only helicases.