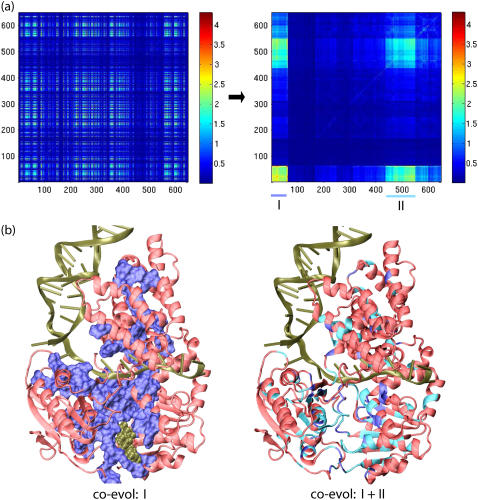
FIGURE 7.
Coevolution analysis of essential residues in helicases related to PcrA. Coevolutionary SCA studies pairwise mutational correlation in protein sequences; results indicate the essential amino acids of proteins (28,29). For the purpose of coevolutionary analysis, one needs to involve large enough protein families, as strictly conserved amino acids (in small families) obviously do not exhibit coevolution. Accordingly, SCA was based on alignment of over 800 sequences of helicases (alignment 3, including PcrA, Rep, UvrD, UvrB, RecQ, Rad54, and NS3, i.e., helicases that share structurally similar domains 1A and 2A). (a) Shown on the left is the correlation map colored according to the SCA correlation matrix elements that describe the mutational coupling strength between two residues in the protein sequence alignment. This correlation map is rearranged on the right, employing a procedure that clusters highly correlated residues (29); two such clustered regions arise and are labeled I and II. (b) Regions I and II are illustrated in the PcrA-DNA complex, shown in cartoon presentation, with the bound ATP in vdW presentation. On the left, coevolutionary region I, the highest autocorrelated region, is displayed in blue surface representation, with red showing the rest of the regions; on the right, both coevolutionary regions, I (blue) and II (light blue), are highlighted. Coevolutionary regions I and II represent the functional core of PcrA-related helicases.