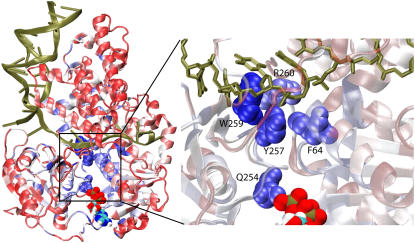
FIGURE 8.
Residues that coevolved with Q254. By means of coevolutionary SCA, we determined residues that mutated jointly with Q254, implying that they are functionally (or structurally) related to this residue. Q254 was singled out for this analysis since it is in close contact with the γ-phosphate of ATP in the substrate state and has been proposed as a Pi sensor coupling the ATP hydrolysis to ssDNA binding (12,50). Shown on the left is the PcrA-DNA complex in cartoon presentation with bound ATP in a vdW presentation. The protein is colored according to the degree of correlation of its residues with respect to coevolution with Q254; this correlation is determined through the SCA correlation matrix (Fig. 7); red regions exhibit low correlation with Q254, whereas blue regions exhibit high correlation. On the right is an enlarged view of the boxed loop region in a, connecting Q254 in the ATP binding pocket to the ssDNA binding interface near R260, with the ssDNA nucleotides in licorice format and key amino acids in vdW format. Note that residues Y257, W259, R260, and F64, which are highly correlated with Q254 in “coevolution”, play important roles in ssDNA binding (8,11,50,53); in particular, the side chains of Y257 and F64, in the product state (no ATP/ADP), form a pocket that accommodates a base from the ssDNA (see nucleotide 18 in Fig. 2 b).