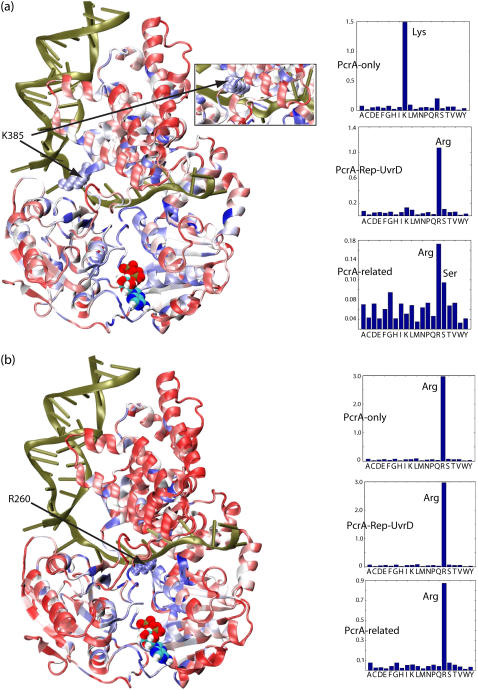
FIGURE 9.
Residues that coevolved with K385 and R260 and their evolutionary substitutions. We have carried out the same evolutionary analysis for K385 and R260 in PcrA–BACST as for Q254 in Fig. 8. We also determined, through sequence alignment, which amino acids are found to replace K385 and R260. Residues K385 and R260 were singled out, since they were identified in our previous study (11) as residues important for the translocation of PcrA. (a) At left is the PcrA-DNA complex in cartoon format with bound ATP in vdW format. The protein is colored according to the degree of correlation of its residues with respect to coevolution with K385: red regions exhibit low correlation with K385, whereas blue regions exhibit high correlation. (Inset) The conformation of K385 in the product state, which differs from that in the substrate state by a 180° rotation toward the ssDNA. At right are the distributions of amino acids found to replace residue 385 in the multiple sequence alignment of PcrA-only helicases (top); PcrA-Rep-UvrD helicases (middle); and PcrA-related helicases (bottom), including not only PcrA, Rep, and UvrD, but also UvrB, RecQ, Rad54, and NS3. Note that K385 is substituted by serine in Rep helicase and arginine in UvrD helicase. (b) Shown on the left is the PcrA-DNA complex with the protein colored according to the degree of correlation of its residues with respect to coevolution with R260. Shown on the right are the distributions of amino acids found to replace residue 260 in the three multiple sequence alignments, as in a. Note that R260 is always relatively conserved as arginine.