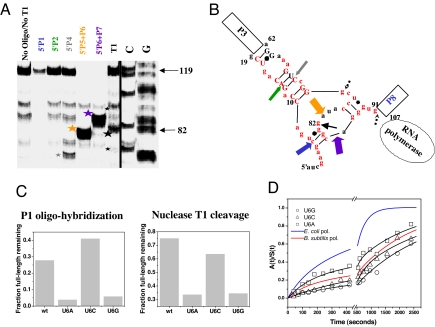
Fig. 2.
Structural analysis of the nascent RNA in the 119-paused complex. (A) Structural mapping of 5′ 32P-labeled 119-paused complex by oligohybridization by using probes complementary to different regions in P RNA and by partial nuclease T1 digestion. The cleavage products are indicated by stars. (B) Proposed structure of the nascent RNA in the 119-paused complex. Colored block arrows show the approximate location and relative intensity of RNase H cleavage upon oligohybridization with the color matching the oligo probe used in A. A black arrow shows the location of the major T1 cleavage product (large star in A). Minor T1 cleavage products are dashed (small stars in A). Red nucleotides indicate the 5′ nucleotides of the long-range helices P1, P2, P4, P5, P6, and P7. Phylogenetically supported base pairs are shown in capital letters. (C) Structural mapping of U6 mutants by T1 nuclease cleavage and by P1 oligohybridization. Mapping with the P2, P4, P5-P6, and P6-P7 oligoprobes produced results similar to wild type (not shown). (D) Folding of U6 mutants when transcribed by E. coli polymerase.