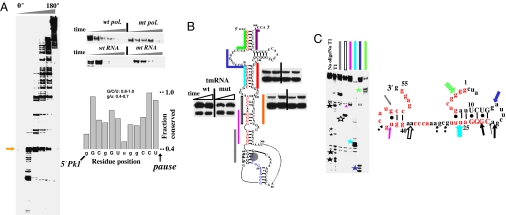
Fig. 4.
Pausing, folding, and structural analysis of E. coli tmRNA. (A Left and Upper Right) Many pause sites are present during transcription by wild-type polymerase. The first major pause (arrow) at U65 is less pronounced in the transcription by the mutant polymerase or through mutation of A51-U65 to U51-A65. Many pause sites are present in the pseudoknot region (residues 200–280). (Lower Right) Sequence preceding the U65 pause is conserved among γ-proteobacteria (manual alignment of 69 sequences). Nucleotides exhibiting especially high levels of conservation are capitalized. (B) Folding monitored by oligohybridization during transcription of wild-type and U25-A82 pausing deficient tmRNA by wild-type polymerase. Upstream portions of the long-range helices are in red and the U65 pause site is shaded. The relative protection from 15–30 seconds is shown. (C Left) Structural mapping of the body-labeled U65 paused complex by oligohybridization and partial nuclease T1 digestion. The six oligos used are complementary to the upstream portions of the long-range helices (matching colors as those in B). The cleavage products are indicated by stars. (Right) Proposed structure of the nascent RNA in paused complex. Phylogenetically supported base pairs are shown in capital letters. Colored block arrows, relative intensity and location of RNase H/oligohybridization with color matching the oligo probe. Black arrows, location of the major T1 cleavage (large stars in gel on the left) with dashed arrows indicating minor cleavage products (small stars in gels on the left).